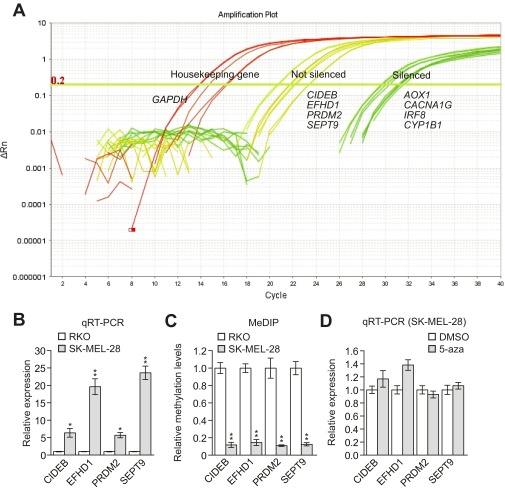
Fig. S2.
CIDEB, EFHD1, PRDM2, and SEPT9 are not hypermethylated and silenced in SK-MEL-28 cells. (A) Amplification plot showing Ct values for silenced genes, nonsilenced genes, and (as a control) a housekeeping gene in SK-MEL-28 cells. The results show that the Ct values for CIDEB, EFHD1, PRDM2, and SEPT9 were 20–22, whereas the Ct values of the other derepressed genes were 28–32, suggesting the four genes were not silenced in SK-MEL-28 cells. ΔRn = Rn+ − Rn−, where Rn is the fluorescence emission intensity of the reporter dye divided by the fluorescence emission intensity of the passive reference dye (Rn+ is the value of the reaction containing all components and Rn− is the value of an unreacted sample). (B) qRT-PCR analysis monitoring expression of CIDEB, EFHD1, PRDM2, and SEPT9 in SK-MEL-28 cells relative to expression observed in RKO cells. The results in SK-MEL-28 cells were normalized to the results obtained in RKO cells, which were set to 1. (C) qRT-PCR analysis monitoring expression of CIDEB, EFHD1, PRDM2, and SEPT9 in SK-MEL-28 cells treated in the presence or absence of the DNA methyltransferase inhibitor 5-azacyticine (5-aza). The results show that 5-aza treatment had no effect on expression of the four genes in SK-MEL-28 cells. (D) Methylated DNA immunoprecipitation (MeDIP) analysis of CIDEB, EFHD1, PRDM2, and SEPT9 in RKO and SK-MEL-28 cells. Methylation was normalized to normalization obtained in RKO cells, which was set to 1. Data are represented as mean ± SD. *P ≤ 0.05; **P ≤ 0.01.