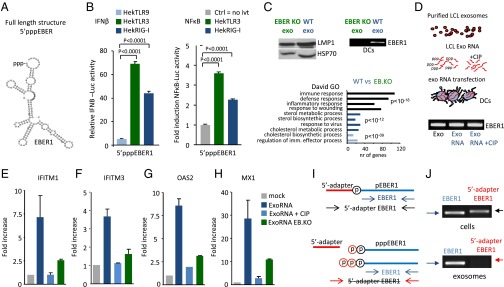
Fig. 4.
The 5′triphosphate moiety of EBER1 is required for antiviral response activation. (A) Predicted secondary structure of EBER1 (RNAfold web server). (B) IFN-β and NF-κB promoter activation (assessed with luciferase reporters) in response to 5′pppEBER1 (ivt) transfection in Hek293 cells expressing TLR9, TLR3, or RIG-I (cells were “primed” with rhIFN-α). Firefly luciferase activity is normalized to Gaussia luciferase activity and data are expressed relative to control. (C) Western blot analysis of LMP1 and Hsp70 in exosomes isolated from LCLs infected with EBER-deficient mutant (EB.KO) or wild-type (wt) EBV (Upper Left). DCs were incubated with EBER1-deficient or EBER1-containing exosomes and the EBER1 levels were assessed by qPCR (Upper Right). Microarray analysis (Lower) shows that antiviral genes are strongly overrepresented in DCs exposed to wild-type exosomes compared with DCs incubated with EBER1-deficient exosomes. (D–H) Exosomal RNA was incubated with CIP to remove the phosphate moieties, and DCs were transfected with CIP-treated or untreated exosomal RNA. EBER1 levels were comparable in all experimental conditions (D). qPCR analysis shows that treatment with CIP completely abrogates the ability of exosomal RNA to induce ISGs. Transcript levels are normalized to GAPDH and expressed as fold-increase relative to mock control (E–H). (I and J) LCL cellular and exosomal RNA were incubated with adapters that can only ligate 5′monophosphate RNAs; qPCR was performed using primers specific for the 5′adapter and 3′EBER1 (I). Full-length EBER1, but not 5′adapter ligated-EBER1, is detectable in LCL exosomes (J).