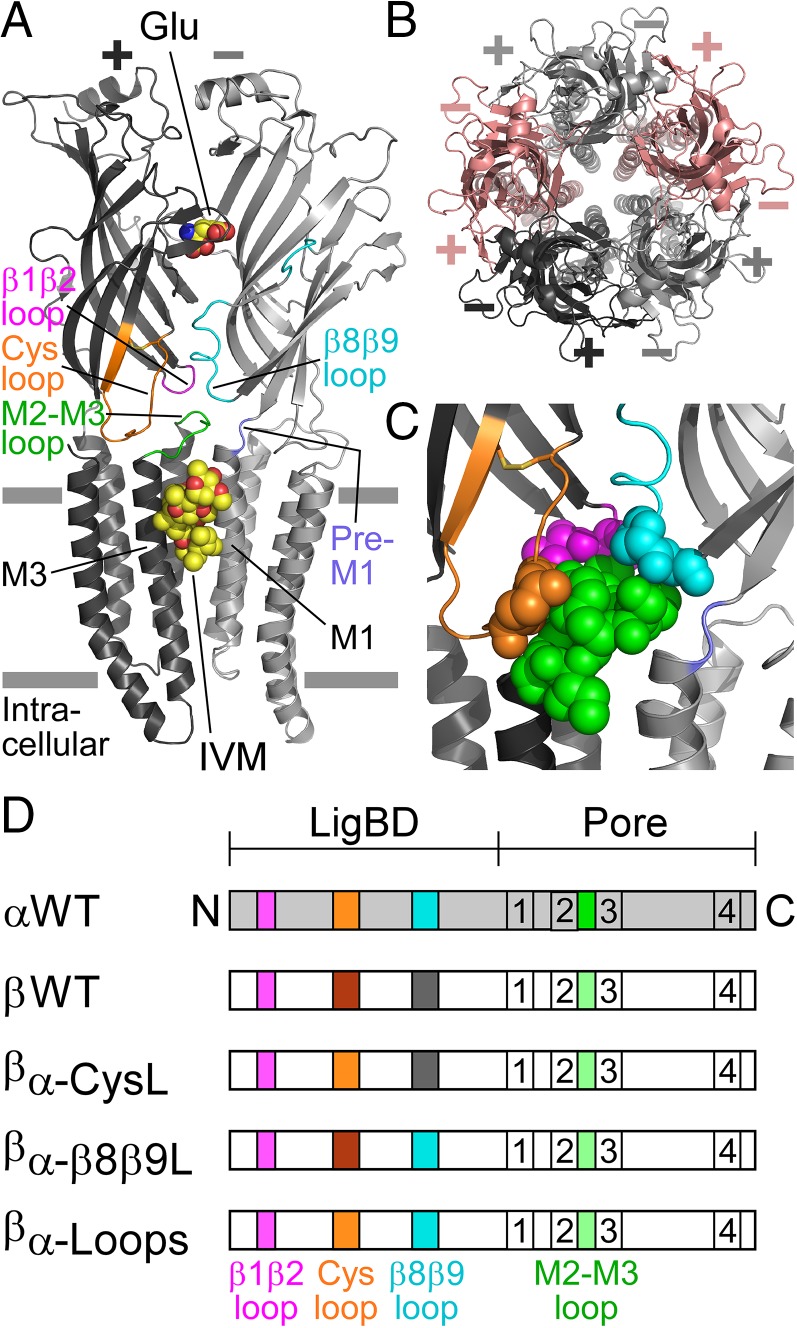
Fig. 1.
Structural characteristics of GluClRs. (A) Two neighboring subunits of the homopentameric GluClαcrystR [Protein Data Bank (PDB) ID code 3RIF] are shown from the side in light and dark gray colors. Wide gray horizontal lines mark the putative membrane borders. The four coupling loops are colored as shown in C and the upper row of D. Glu and IVM are shown as space-filling models with carbon, oxygen, and nitrogen atoms colored in yellow, red, and blue, respectively. They are bound at the α(+)/α(−) intersubunit interface far away from each other: Glu in the extracellular LigBD and IVM in the upper part of the pore-domain periphery, between M1 (of the light gray subunit) and M3 (of the dark gray subunit). Note that in Cys-loop receptors, the principal and complementary faces of a neurotransmitter-binding pocket are formed by the (+) and (−) sides of two adjacent subunits, respectively. (B) Top view of the GluClαcryst pentamer showing five identical subunits, which are colored differently to highlight the intersubunit interfaces located between the (+) and (−) sides. (C) Space-filling models of residues belonging to the coupling loops, which create an extensive bond network at the interface between the LigBD and the ion-channel pore domain. (D) Schemes of GluClR subunits used in this study. The M1–M4 transmembrane segments are numbered 1–4. Different colors reflect differences in amino acid sequences (Fig. S1).