Fig. 4.
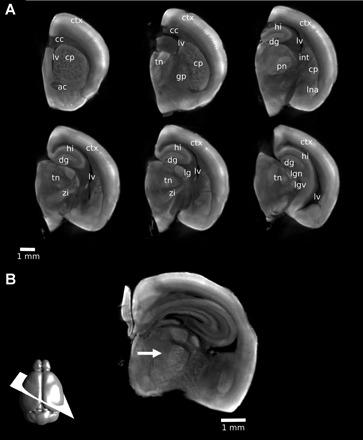
Visualizing neuroanatomy with autofluorescence. With a 1% PFA fixation, a number of structures can be identified in the mouse brain. Two-dimensional (2D) slices were extracted from a three-dimensional (3D) OPT image of a single hemisphere. Structures identified are labeled on multiple coronal slices (A) to indicate the 3D nature of the dataset: ctx, cortex; cc, corpus callosum; lv, lateral ventricle; cp, caudate/putamen; ac, anterior commissure; tn, thalamic nuclei; gp, globus pallidus; dg, dentate gyrus; hi, hippocampus; int, internal capsule; pn, posteriomedial nucleus; lna, lateral nucleus of the amygdala; zi, zona incerta; lg, lateral geniculate nucleus; lgd, dorsal lateral geniculate nucleus; lgv, ventral lateral geniculate nucleus. With 3D imaging, 2D slices at arbitrary orientations can be generated retrospectively. In B, the thalamic barreloids are visible on a 2D oblique slice through a 3D OPT image of a 3 mm section of the brain (arrow). The orientation of the slice is shown schematically at the bottom left.
