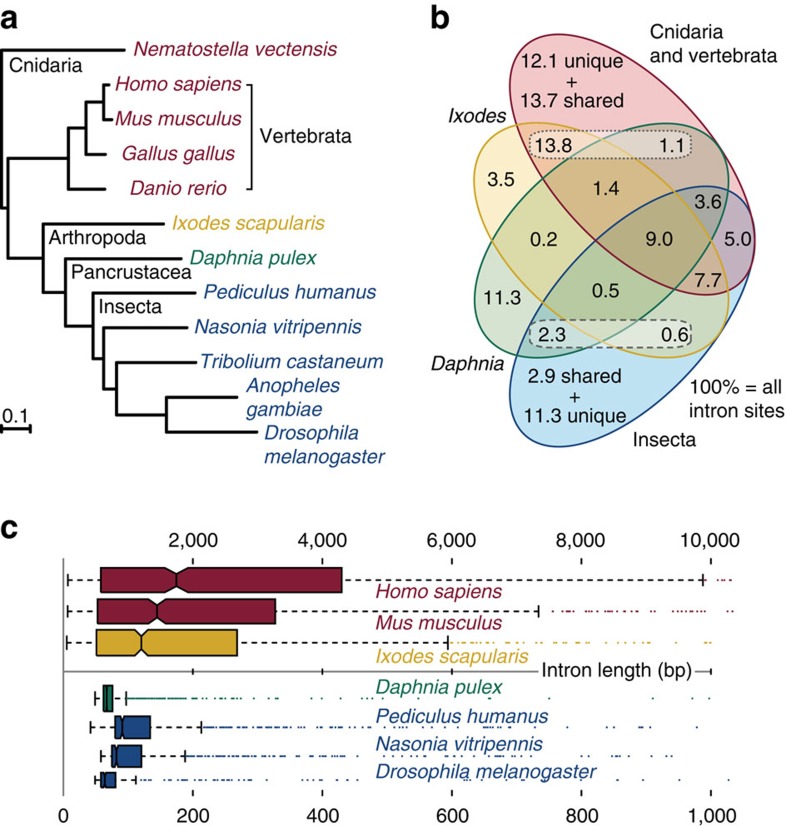
Figure 3. Molecular and intron evolution of Ixodes scapularis orthologs.
(a) The species phylogeny computed from the concatenated alignment of single-copy orthologous protein-coding genes confirms the position of the Subphylum Chelicerata at the base of the arthropod radiation, an outgroup to the clade Pancrustacea that contains crustaceans and hexapods. The average rate of I. scapularis molecular evolution is slower than that in the fast-evolving dipterans (Anopheles gambiae and Drosophila melanogaster), comparable to other representative arthropods for which genome sequences are available, and faster than that of vertebrates. (b) Quantification of the proportions of shared and unique intron positions from well-aligned regions of universal orthologs reveals that, compared with the crustacean, Daphnia pulex, I. scapularis shares more than 10 times as many introns exclusively with at least one of the five outgroup species (from Cnidaria and Vertebrata) (dotted box, 13.8% versus 1.1%). Conversely, D. pulex has more intron positions exclusively in common with the representative insects (dashed box, 2.3% versus 0.6%). (c) I. scapularis intron lengths are more similar to those of introns from orthologous genes in the vertebrates Homo sapiens and Mus musculus, and are an order of magnitude longer than introns from the pancrustacean species analysed. The intron length distributions are shown for ancient introns found in both I. scapularis and D. pulex and at least one of the five outgroup species and at least one insect; boxplots indicate medians, first and third quartiles, and whiskers.