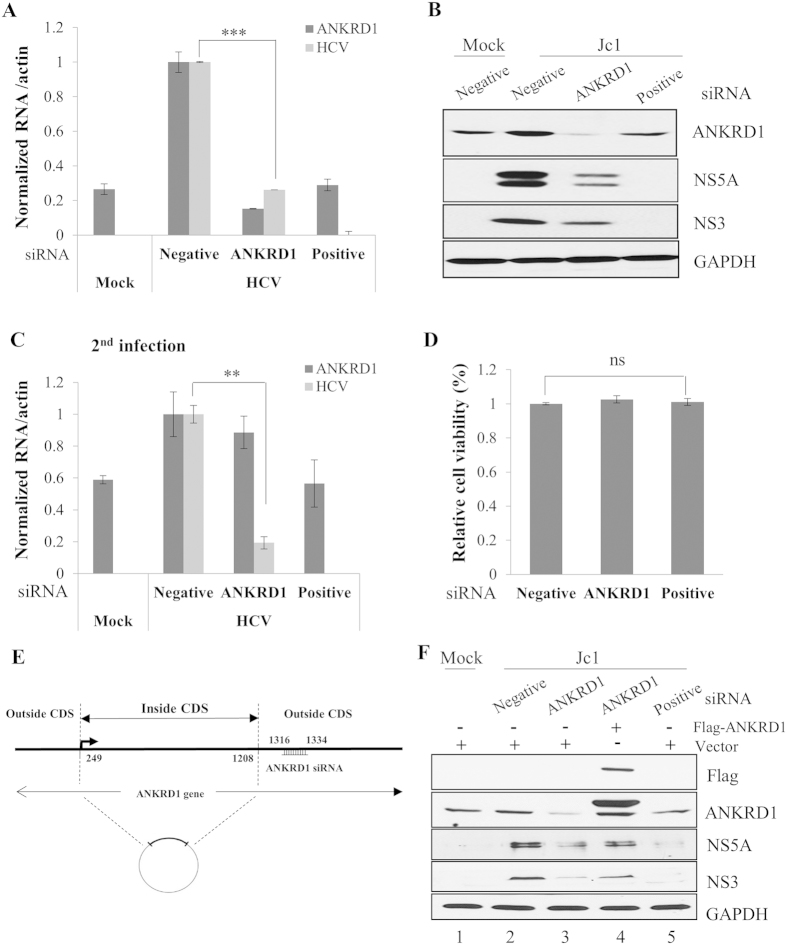
Figure 5. ANKRD1 is required for HCV propagation.
(A) Huh7.5 cells were transfected with 20 nM of the indicated siRNAs for 48 h and then either mock infected or infected with Jc1 for 4 h. At 2 days postinfection, intracellular RNA levels were determined by qRT-PCR. Data represent averages from at least three independent experiments. The asterisks indicate significant differences (**P < 0.01; ***P < 0.001). (B) Huh7.5 cells were transfected with 20 nM of the indicated siRNAs for 48 h and then either mock infected or infected with Jc1 for 4 h. At 2 days postinfection, total cell lysates were immunoblotted with the indicated antibodies. Positive indicates HCV-specific siRNA targeting the 5′ nontranslated region (NTR) of Jc1. Negative depicts universal negative-control siRNA. (C) Naïve Huh7.5 cells were infected with Jc1 harvested from (B) for 4 h. At 48 h postinfection, intracellular HCV RNA levels were analyzed by qRT-PCR to determine infectivity. The asterisks indicate significant differences (**P < 0.01). (D) Huh7.5 cells were transfected with 20 nM of the indicated siRNAs. At 96 h after transfection, cell viability was determined by MTT assay. (E) Schematic diagram showing the siRNA target site outside of ANKRD1 coding sequence (CDS). (F) Huh7.5 cells were transfected with 20 nM of the indicated siRNAs for 24 h. The cells were further transfected with either vector or Flag-tagged ANKRD1 plasmid for 24 h. Cells were then either mock infected or infected with Jc1. At 48 h postinfection, total cell lysates were immunoblotted with the indicated antibodies.