FIGURE 1.
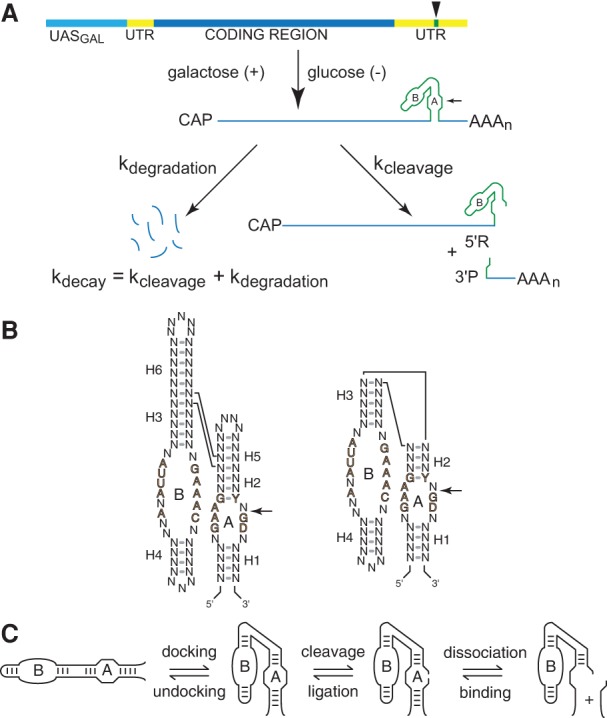
Chimeric ribozyme mRNAs as reporters of intracellular RNA folding. (A) Quantitative analysis of RNA folding in yeast. Ribozyme sequences (green) are inserted into the 3′ UTR (yellow) of the yeast PGK1 gene and transcribed under the control of the GAL1-10 upstream activation sequence, UASGAL (aqua), to allow measurement of chimeric mRNA decay kinetics after glucose inhibition. Self-cleaving mRNA decays both through self-cleavage (kcleavage) and through the normal mRNA degradation pathway (kdegradation), so self-cleavage accelerates decay by an amount that corresponds to the intracellular cleavage rate. (B) The two forms of hairpin ribozymes. Natural ribozymes (left) consist of two helix-loop-helix elements that assemble in the context of a four-way helical junction, whereas minimal ribozymes (right) assemble in the context of a two-way helical junction. Reversible cleavage of the reactive phosphodiester bond in Loop A is indicated by arrows. Defined nucleotides represent conserved sequences important for catalytic activity. (C) The hairpin ribozyme self-cleavage pathway. The minimal hairpin ribozyme (left) has two helix-loop-helix domains, A and B, that dock to form the active site, which catalyzes reversible cleavage of a specific phosphodiester bond in Loop A. Cleavage product dissociation occurs through unwinding of the intermolecular H1 helix. Diagrams adapted from Watson and Fedor (2009) and Mahen et al. (2010).
