Figure 4. Perlecan Dm IV-3 cleavage patterns by MMP-7.
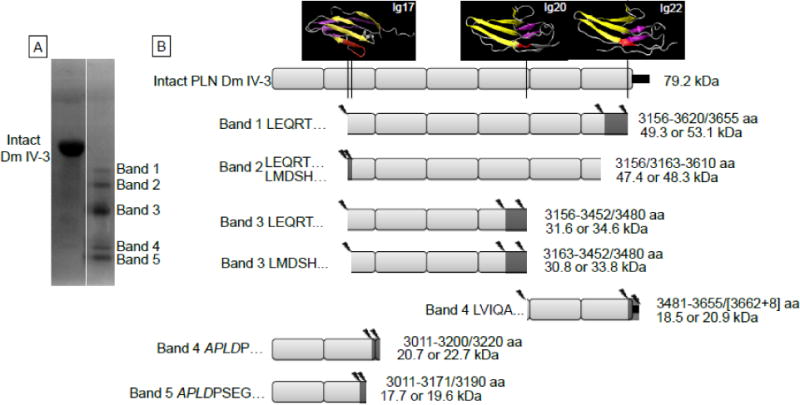
A) The Coomassie stained PVDF membrane of Dm IV-3 either alone (intact) or incubated with MMP-7. The five primary protein bands were analyzed for N-terminal sequencing by Edman degradation. B) Likely cleavage patterns for each band of MMP-7 digested Dm IV-3 given the sequenced N-termini, MALDI mass measurements and LC-MS/MS identified peptides. The left side of each indicated fragment identifies the band N-terminus sequencing found, and the right indicates the likely C-terminus amino acids (amino acid, aa) MMP-7 cleavage site and the resultant apparent molecular weight (kDa) that corresponds approximately to the masses found by MALDI and in the Coomassie stained PVDF. Above intact Dm IV-3 are the PHYRE modeled Ig modules with the indicated MMP-7 cleavage sites shown in red. Cleavage mostly occurs at amino acids in the linking region between Ig modules or in the middle of the Ig module linking the interfacing beta strands. Additionally, each MMP-7 cleavage site produces an aliphatic N-terminus such as leucine, isoleucine, or valine. The small black rectangle at the end of the Dm IV-3 schematic indicates the V5 epitope and polyhistidine tag. The asterisks indicate the 3135 polyclonal antibody binding site on Dm IV-3.
