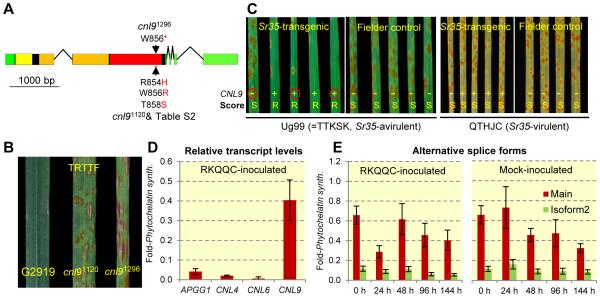
Fig. 2.
(A) CNL9 gene structure. Green=UTR, black= coding exons, yellow= coiled-coil domain, orange= nucleotide-binding domain, red= leucine-rich repeat domain, arrows= amino acid changes in susceptible induced mutants cnl91296 (W856*) and cnl91120 or natural mutants (Table S2, RLWFT to HLRFS) (B) Infection types produced on T. monococcum G2919 and CNL9 mutants cnl91120 and cnl91296 inoculated with Pgt race TRTTF. TRTTF is Sr35-avirulent and Sr21-virulent, which is required because of the presence of Sr21 in these lines. (C) Infection types on seedlings of T1 lines from event #1123 segregating for the CNL9 transgene. Plants carrying the CNL9 transgene (+) were resistant to Ug99 (R) and plants without the transgene (−) were susceptible (S) (Table S8). When inoculated with Sr35-virulent race QTHJC all plants were susceptible suggesting similar race specificity between the transgenic and the natural Sr35. Red circles indicate available progeny tests in Figure S5. (D) Relative transcript levels of candidate genes APPG1, CNL4, CNL6, and CNL9 (main isoform) in G2919 six days after inoculation with race RKQQC (E) Transcript levels of CNL9 main isoform (red) and isoform two (green, retained intron) in mock- or race RKQQC inoculated G2919 plants. Leaves were collected at 0, 24, 48, 96 and 144 h after inoculation. Transcript levels are expressed relative to the Phytochelatin synthase internal control using the 2−ΔCt method. Bars are standard errors of the means based on six biological and two technical replicates.