Abstract
RNA viruses exploit host cells by co-opting host factors and lipids and escaping host antiviral responses. Previous genome-wide screens with Tomato bushy stunt virus (TBSV) in the model host yeast have identified 18 cellular genes that are part of the actin network. In this paper, we show that the p33 viral replication factor interacts with the cellular cofilin (Cof1p), which is an actin depolymerization factor. Using temperature-sensitive (ts) Cof1p or actin (Act1p) mutants at a semi-permissive temperature, we find an increased level of TBSV RNA accumulation in yeast cells and elevated in vitro activity of the tombusvirus replicase. We show that the large p33 containing replication organelle-like structures are located in the close vicinity of actin patches in yeast cells or around actin cable hubs in infected plant cells. Therefore, the actin filaments could be involved in VRC assembly and the formation of large viral replication compartments containing many individual VRCs. Moreover, we show that the actin network affects the recruitment of viral and cellular components, including oxysterol binding proteins and VAP proteins to form membrane contact sites for efficient transfer of sterols to the sites of replication. Altogether, the emerging picture is that TBSV, via direct interaction between the p33 replication protein and Cof1p, controls cofilin activities to obstruct the dynamic actin network that leads to efficient subversion of cellular factors for pro-viral functions. In summary, the discovery that TBSV interacts with cellular cofilin and blocks the severing of existing filaments and the formation of new actin filaments in infected cells opens a new window to unravel the way by which viruses could subvert/co-opt cellular proteins and lipids. By regulating the functions of cofilin and the actin network, which are central nodes in cellular pathways, viruses could gain supremacy in subversion of cellular factors for pro-viral functions.
Author Summary
The actin network, which is a central node in cellular pathways, is frequently targeted by various pathogens to modulate cellular responses. In this paper, the authors show that TBSV interacts with cofilin actin depolymerization factor leading to inhibition of the dynamic function of the actin network in infected cells. This allows TBSV to utilize the existing actin filaments to efficiently recruit host proteins and lipids for viral replication and to build viral replication compartments for robust viral replication. Altogether, subversion of the actin network by TBSV is a key step for the virus to gain access to cellular resources required for virus replication.
Introduction
Plus-stranded (+)RNA viruses, which are important pathogens of plants, animals and humans, co-opt a number of host-coded proteins and lipids to facilitate the replication process [1–6]. These viruses also remodel host membranes and alter host cellular pathways to take advantage of host resources and to avoid recognition by host antiviral defenses. Characterization of an increasing number of host factors involved in (+)RNA virus replication has already revealed intriguing and complex interactions between various viruses and their hosts. Functional studies with selected host proteins have revealed a plethora of activities preformed by these host proteins during RNA virus infections [1,3,7–11]. In spite of the intensive efforts, our current cataloging of host factors is still far from complete and our current knowledge on the role of the identified host factors is incomplete.
One of the advanced viral systems to study virus-host interactions is Tomato bushy stunt virus (TBSV), a small (+)RNA virus, which can replicate in the model host yeast (Saccharomyces cerevisiae) [12–16]. TBSV replication requires two viral-coded proteins, namely the p33 and p92pol replication proteins. Albeit these proteins have overlapping sequences, they have different functions. p33, which has RNA chaperone activity, has been shown to recruit the TBSV (+)RNA to the cytosolic surface of peroxisomal membranes, the sites of replication [17–20]. The p92pol has RNA-dependent RNA polymerase (RdRp) activity and binds to p33 to assemble the membrane-bound functional viral replicase complex (VRC) [14,19,21–24].
The activities of TBSV replication proteins, however, are affected by numerous host proteins [3,7–9,25]. Indeed, over 500 host genes/proteins that affected TBSV replication and/or recombination, have already been identified by using multiple genome-wide screens of yeast and global proteomics approaches [16,26–31]. Moreover, the tombusvirus VRC contains several host proteins [32–34], including heat shock protein 70 (Hsp70), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), eukaryotic elongation factor 1A (eEF1A), eEF1Bγ, the DDX3-like Ded1, eIF4AIII-like RH2 and DDX5-like RH5 DEAD-box RNA helicases, and the ESCRT (endosomal sorting complexes required for transport) family of host proteins [25,33,35–39]. These proteins are required for VRC assembly or affect viral RNA synthesis [3,35,38,40–42]. The TBSV replication process also depends on phospholipids and sterols, which are actively recruited to the site of viral replication [43–47].
Previous genomic and proteomic screens have revealed that Cof1p (cofilin in mammals) interacts with the tombusvirus p33 replication protein, and a mutation in COF1 enhances TBSV RNA replication in yeast cells [30,48], suggesting that COF1 could be an important host restriction factor. Cof1p is a major modulator of actin filament disassembly and an essential protein for yeast growth [49,50]. The major cellular function of Cof1p is to preferentially bind to ADP-actin subunits in actin filaments that results in twisting and severing the actin filaments [51,52]. Actin filament disassembly via Cof1p-induced depolymerization is required for remodeling of the actin cytoskeleton by providing free actin monomers as substrates for new filament formation [53–55]. Whether cofilin facilitates actin filament assembly or disassembly depends on the concentration of cofilin relative to actin. Overall, cofilins are conserved in eukaryotic cells and are essential from yeast to humans. Cofilins are known to affect many cellular pathways, including cell motility, cytokinesis, endocytosis, receptor functions, apoptosis, phospholipid metabolism, oxidative stress and gene expression via acting as a chaperone for nuclear actin [53,54,56,57]. Cofilins are also involved in several diseases, such as Alzheimer’s disease and ischemic kidney disease and other pathophysiological defects, such as infertility, immune deficiencies, inflammation, cancer, cognitive impairment [54,56,57].
Actin is highly conserved and abundant protein that exists in two forms in cells: globular monomeric (G-actin) and the active filamentous polymeric (F-actin) form. Actin undergoes multiple cycles of rapid nucleation, polymerization and disassembly, which is needed for remodeling the actin cytoskeleton. This organization of actin filaments is needed for vesicle transport, endocytosis, cell division and other functions in response to stimuli [58]. The cell frequently remodels the actin cytoskeleton with the help of ~100 highly conserved accessory proteins [58]. Regulation and activation of the accessory proteins, such as Arp2p and Arp3p actin nucleators and Cof1p, is needed to organize actin filaments in specific regions of the cytoplasm to carry out various functions.
Viruses dramatically reorganize the infected cells to support viral replication. Since the actin network is a key element in cell organization and transport of cellular proteins, lipids and intracellular organelles, viruses might target the actin network to reprogram cellular pathways and aid VRC assembly [59].
In this work, we show that cofilin actin depolymerization factor is targeted by tombusviruses to promote virus replication. Using temperature-sensitive (ts) Cof1p mutants at the semi-permissive temperature, we find increased level of TBSV RNA accumulation in yeast cells. The in vitro activity of the tombusvirus replicase prepared from cof1-8 ts yeast was also higher than the activity of the replicase prepared from wt yeast. Also, over-expression of Cof1p or a plant homolog (Adf2) reduced TBSV accumulation in yeast. In addition, we show that TBSV replication depends on actin organization. Accordingly, inhibition of actin dynamic function via act1 mutations in yeast or pharmacological inhibitors in plant cells led to increased TBSV replication. We demonstrate that the actin network affects the ability of TBSV to recruit host proteins and sterols to the viral replication sites. Altogether, the emerging picture is that TBSV, via direct interaction between p33 and Cof1p, controls cofilin activities to obstruct the dynamic actin network to lead to efficient subversion of cellular proteins and sterols for pro-viral functions.
Results
Cof1p is a regulator of TBSV replication
In our previous high throughput screens, we observed that the actin network was represented by one of the highest number of genes identified for TBSV (namely 18 genes). Particularly, we were interested in COF1, since Cof1p has been shown to interact with the tombusviral p33 replication protein [30]. Since Cof1p is a key actin depolymerization factor [54,55], binding of the tombusviral p33 replication protein to Cof1p might inhibit Cof1p interaction with ADP-actin and block actin disassembly and recycling of actin monomers to form new actin filaments.
To test if the interaction between Cof1p and p33 replication protein is relevant during TBSV replication, we used a temperature-sensitive (ts) Cof1p mutant yeast in viral replication studies. Interestingly, the data from Northern blot analysis showed that cof1-8 ts yeast supported TBSV replicon (rep)RNA accumulation at ~8-fold increased level at the semi-permissive temperature in comparison with yeast carrying wt COF1, while the two strains showed comparable TBSV repRNA accumulation at the permissive temperature (compare lanes 7–12 versus 1–6, Fig 1A and 1B). These data indicate that Cof1p is an inhibitor of TBSV RNA replication in yeast host. The amount of p33 and p92pol was comparable in these yeast strains at the semi-permissive temperature, suggesting that Cof1p does not affect translation or stability of the viral replication proteins. Using the cof1-5 ts mutant of Cof1p also showed ~3.5-fold increased TBSV repRNA accumulation at the semi-permissive temperatures (S1A Fig), further supporting that Cof1p affects TBSV replication.
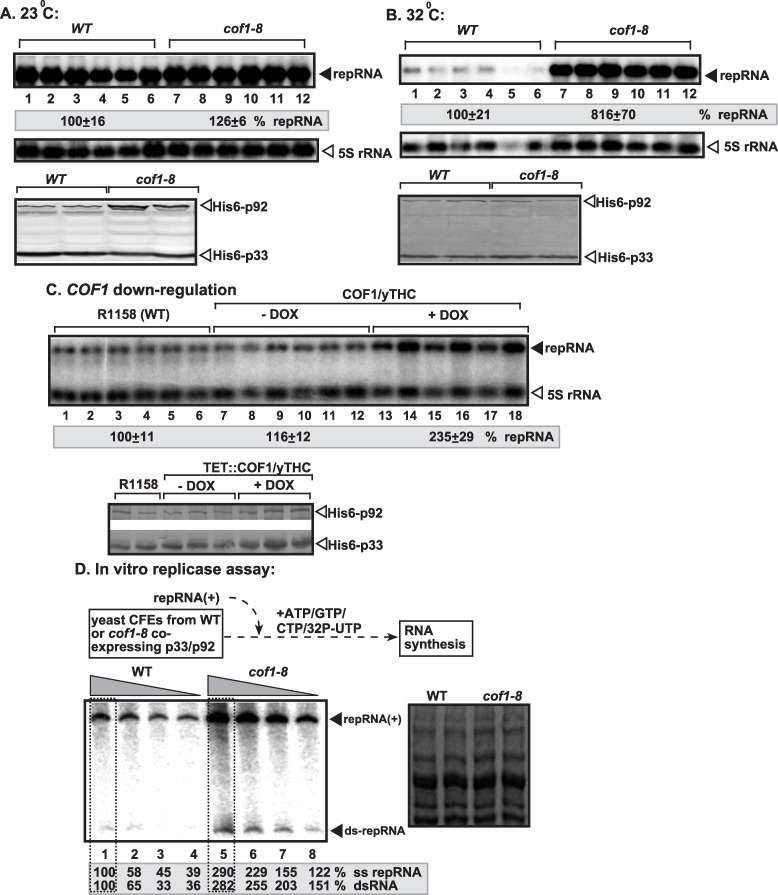
Fig 1. Cof1p inhibits TBSV repRNA accumulation in yeast.
(A-B) Accumulation of TBSV repRNA in cof1-8 ts or wt yeast at the permissive (23°C, panel A) or semi-permissive (32°C, panel B). To launch TBSV repRNA replication, we expressed His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter in the parental (BY4741) and cof1-8 ts yeast strains. Northern blot analysis was used to detect DI-72(+) repRNA accumulation, which was normalized based on 5S rRNA. Typhoon FLA 9500 system (GE) and ImageQuant TL software were used to detect and quantify the bands in the gels. Each experiment was repeated three times. Bottom two panels: Western blot analysis of the accumulation level of His6-p33 and His6-p92 proteins using anti-His antibody. Throughout the paper, +/- means standard deviation. (C) Depletion of Cof1p level in yeast leads to increased repRNA accumulation. Note that doxycycline (+dox samples) leads to depletion of Cof1p expressed from the regulatable TET promoter. Replication of the TBSV DI-72(+) repRNA in wt and TET::COF1 yeasts co-expressing the tombusvirus p33 and p92 replication proteins was measured by Northern blotting 24 h after initiation of TBSV replication. Each sample is obtained from independent yeast colonies. The experiments were repeated two-to-three times. (D) Top: Scheme of the TBSV in vitro replication assay. The CFEs were prepared from wt or cof1-8 ts yeast expressing the tombusvirus p33 and p92 replication proteins at the semi-permissive temperature. Then, the same amount (1 μg) of DI-72(+) repRNA was added to each reaction, followed by in vitro tombusvirus replication assay. Note that the CFE-based assay contained 4x (lanes 1 and 5), 3x (lanes 2 and 6) 2x (lanes 3 and 7) and 1x (lanes 4 and 8) amounts of CFEs. The non-denaturing PAGE analysis shows the emergence of single-stranded (+)repRNAs and dsRNAs (replication intermediate) as indicated. The image on the right shows the coomassie blue-stained total proteins in CFE preparations after SDS-PAGE analysis to show the presence of comparable amounts of total proteins in the CFEs.
To obtain additional evidence on the inhibitory role of Cof1p, we down-regulated Cof1p level in yeast using the titratable TET promoter [60]. We found that TBSV repRNA accumulation increased by ~2.5-fold when Cof1p was down-regulated (lanes 13–18, Fig 1C). Thus, TBSV replication is stimulated by reduced level of Cof1p in yeast.
To test if Cof1p affects the viral replicase activity, we obtained cell-free extracts (CFE) from cof1-8 ts and wt yeast, co-expressing p33 and p92pol. The CFE was programmed with the TBSV (+)repRNA in vitro, allowing one cycle of full TBSV repRNA replication [21]. We observed ~3-to-4-fold higher repRNA replication in CFE from cof1-8 ts than in CFE from wt yeast (Fig 1D, lanes 5–8 versus 1–4). Interestingly, both the newly made (+)repRNA and double-stranded dsRNA (correlating with the minus-stranded RNA [61]) levels increased to a similar extent in CFE from cof1-8 ts (Fig 1D). Thus, Cof1p affects the early stage of TBSV replication, possibly VRC assembly prior to (-)-strand RNA synthesis. Altogether, the in vitro data are consistent with the results from yeast, supporting that Cof1p is a negative regulator of TBSV replication.
Over-expression of Cof1p inhibits TBSV RNA accumulation
Over-expression of the zz-His6-tagged Cof1p reduced TBSV repRNA accumulation by ~50% in both cof1-8 ts and wt yeast at the permissive temperature (23°C, Fig 2A, lanes 1–4 and 5–8). The inhibitory effect of Cof1p was even more pronounced at the semi-permissive temperature (32°C), resulting in ~30-fold reduction in TBSV repRNA level in cof1-8 ts yeast over-expressing Cof1p (Fig 2B, lanes 1–2 versus 5–6). The amount of p33 and p92pol was comparable in these yeast strains at both temperatures. We also found a strong inhibitory effect on TBSV repRNA accumulation when the native (nontagged) Cof1p was over-expressed in cof1-8 ts and wt yeast strains (S2 Fig).
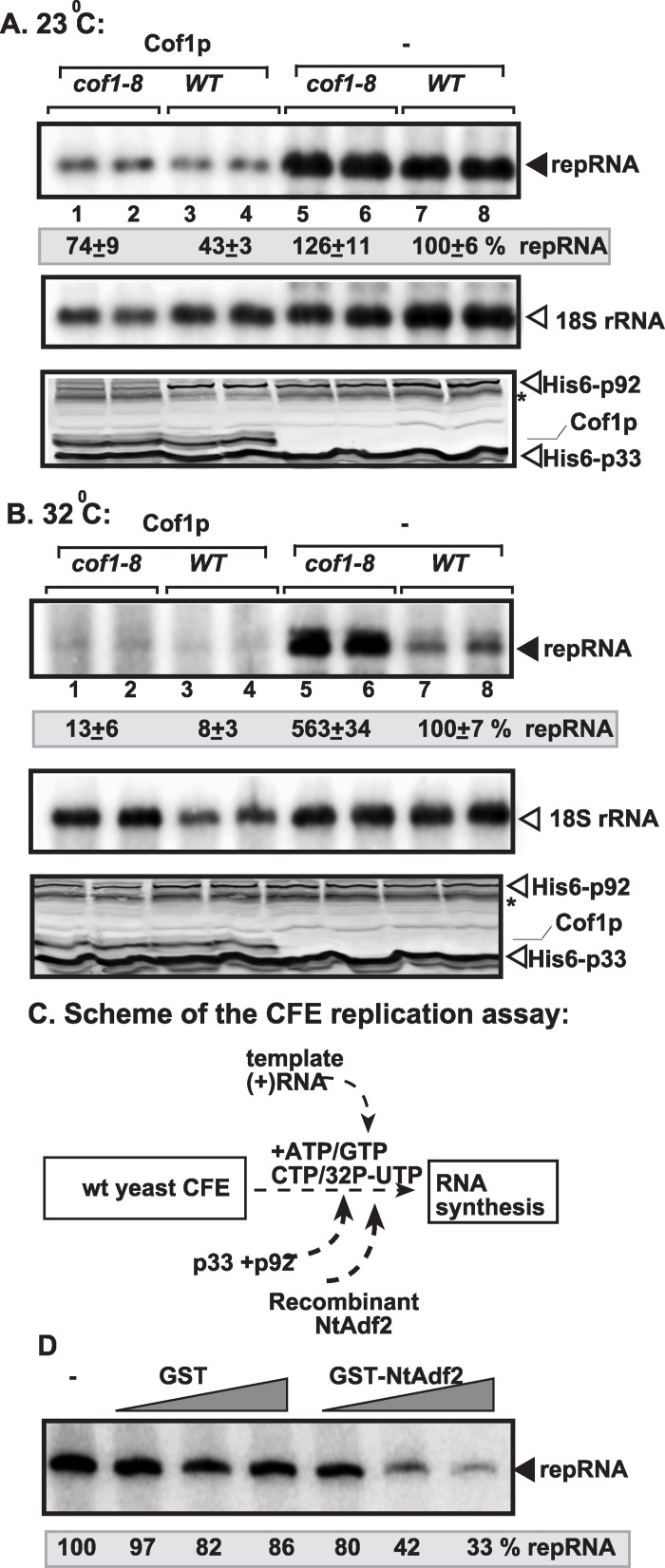
Fig 2. Over-expression of Cof1p inhibits TBSV repRNA accumulation in yeast.
(A-B) The wt or cof1-8 ts yeast co-expressed the His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter. Over-expression of His6-Cof1p was done from the GAL1 promoter. Top images: Northern blot analysis of TBSV repRNA in yeast samples over-expressing Cof1p or without over-expression is shown. repRNA replication took place for 24 hours at 23°C or 32°C in wt or cof1-8 ts yeast before RNA analysis. The accumulation level of DI-72(+) repRNA (shown in percentage) was normalized based on 18S rRNA. Middle panel shows the level of 18S rRNA. Bottom panels: Western blot analysis of the co-expressed 6xHis-tagged proteins with anti-His antibody is shown. (C) Scheme of the CFE-based TBSV replication assay. Purified recombinant p33 and p92pol replication proteins of TBSV and in vitro transcribed TBSV DI-72 (+)repRNA were added to the CFE prepared from wt yeast strain. The affinity-purified recombinant Nicotiana tabacum Adf2 (Cof1 homolog) was added before the CFE-based replication assay as shown. (D) Inhibition of TBSV replication by recombinant NtAdf2 in vitro. Denaturing PAGE analysis of the 32P-labeled TBSV repRNA products obtained in the CFE-based TBSV replication assay in the presence of recombinant GST-NtAdf2p (1x = 200 ng) as shown. Each experiment was repeated three times.
To test if cofilin can directly inhibit TBSV replication, we purified recombinant Adf2 of Nicotiana tabacum, a plant homolog of the yeast Cof1p [62–64]. Adding purified recombinant NtAdf2 to yeast CFE (as shown schematically in Fig 2C) performing one full cycle of TBSV replication (including both minus- and plus-strand synthesis) resulted in up to 70% inhibition of TBSV RNA synthesis (Fig 2D). This experiment showed that cofilin/ADF could directly inhibit TBSV replication in vitro, likely due to sequestering the p33 replication protein.
Formation of large tombusvirus replication compartments in cof1-8ts yeast
To test if the higher level of TBSV RNA replication and the more active tombusvirus replicase from cof1-8 ts yeast are due to the different subcellular localization or an altered distribution of the replication proteins, we used confocal laser microscopy imaging of live yeast cells. This study revealed that p33 replication protein formed a few large punctate structures in cof1-8 ts yeast at the semi-permissive temperature in contrast with the more numerous, but smaller punctate structures in wt yeast (Fig 3). However, YFP-p33 was co-localized with the peroxisomal marker protein (Pex13p) in cof1-8 ts yeast (Fig 3A), suggesting that p33 is localized to peroxisome-derived membranes in both cof1-8 ts and wt yeast cells. Altogether, the presence of large replication foci in cof1-8 ts yeast suggests that tombusvirus replication proteins are likely more efficient in organizing the viral replication compartments containing many VRCs than in wt yeast cells. The formation of large p33-containing peroxisomal structures in cof1-8 ts yeast expressing the Cof1 ts mutant (Fig 3A) were similar to those observed with phospholipid synthesis mutants that also supported TBSV replication at a high level [44,46,65]. These data suggest efficient assembly of VRCs when cofilin is mutated.
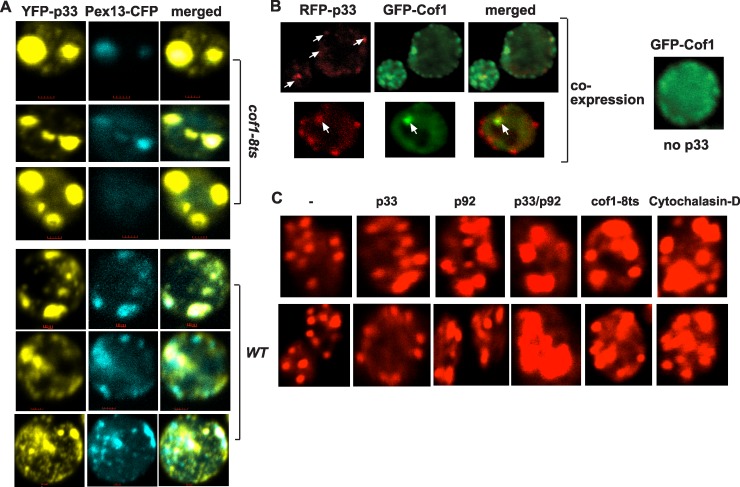
Fig 3. The viral replication proteins inhibit the actin network via interaction with Cof1p.
(A) Confocal laser microscopy images show the co-localization of Pex13-CFP with YFP-p33 expressed from ADH1 promoter in cof1-8 ts or in BY4741 (parental strain) yeast strains at the semi-permissive temperature (32°C). The merged images show in single yeast cells the co-localization of Pex13-CFP with YFP-p33 in fewer but larger punctate structures, which is indicative of large replication organelles, in cof1-8 ts than in wt yeast cells. Note that Pex13 represents a peroxisomal marker, where TBSV forms the VRCs and performs RNA replication. The bars represent 1 μm. (B) Partial co-localization of Cof1p and p33 replication protein in yeast cells. Top panel shows the early time point (5 h), while the bottom panel shows the late time point (24 h) after the induction of protein expression from the GAL1 promoter in cof1-8ts yeast cells. The shift to the semi-permissive temperature (32°C) was done 2 h prior to the induction of Cof1p and p33 expression. Note also the altered localization pattern of GFP-Cof1 in yeast co-expressing p33 in comparison with the more even distribution of GFP-Cof1 in the absence of viral components. The GFP-cofilin construct has S65T-GFP and a 12–amino acid linker inserted between amino acids N74 and G75 of Cof1p. (C) Rhodamine-phalloidin staining of actin in wt yeast expressing p33, p92, or p33+p92 replication proteins in comparison with cof1-8 ts yeast (grown at the semi-permissive 32°C) or wt yeast treated with Cytochalasin-D, an inhibitor of new actin filament formation in yeast. Note that the control (left panels), cof1-8 ts yeast and Cytochalasin-D treated yeasts do not have viral components. The larger size of cortical actin patches are due to the inhibition of actin monomer recycling and lack of dynamic new actin filament formation. Two sets of independent yeast cells are shown in each experiment.
To study if the presence of p33 changes the subcellular localization of Cof1p, we co-expressed RFP-tagged p33 and GFP-tagged Cof1p in cof1-8 ts yeast cells at the semi-permissive temperature, followed by confocal laser microscopy. Interestingly, in the presence of p33, Cof1p showed not only diffused localization in the cytosol, but also formed several punctate structures, which frequently co-localized with p33 (Fig 3B, left images). In contrast, GFP-Cof1p was present mostly in the diffused form in the cytosol in cof1-8 ts yeast in the absence of p33 (Fig 3B, right panel). These results indicate that a fraction of Cof1p is co-localized with p33 at both early (5 hour) and late time points, indicating that the co-expressed p33 can change the subcellular localization of Cof1p in cof1-8 ts yeast. The viral replication protein-induced relocalization of Cof1p might inhibit the normal cellular function of Cof1p (see Discussion).
Since Cof1p is a key actin depolymerization factor [54,55], and binds to the tombusviral p33, it is plausible that p33 might inhibit Cof1p interaction with ADP-actin and block actin filament disassembly and recycling of actin monomers that is needed to form new actin filaments. Indeed, co-expression of p33 and p92 replication proteins in yeast led to the formation of a large actin-filament network (called actin patches, which contain short, branched actin filaments with 1–2 minute turnover time) in the cell, visible under confocal microscope (Fig 3C). Formation of large cortical actin patches is indicative of lack of Cof1-driven disassembly of actin filaments and inhibition of dynamic actin function [52,58]. Indeed, blocking Cof1p function via applying semi-permissive temperature in cof1-8 ts yeast or using the actin polymerization inhibitor, Cytochalasin-D, has led to similar actin organization (large cortical actin patches) to that induced by p33/p92 expression (Fig 3C).
The cortical actin patches were also influenced by expressing either p33 or p92 alone, albeit the effects were lesser than those observed with co-expression of p33 and p92 (Fig 3C). These observations suggest that p33 and p92 interferes with the normal organization of actin in yeast, perturbing actin organization as efficiently as Cof1p mutation or chemical inhibition of actin polymerization. It is worthwhile to mention that p92 shares an identical N-terminal sequence with the full-length p33 and both p33 and p92 can affect actin organization under the experimental conditions (Fig 3C). Based on these observations, we propose that TBSV replication proteins actively block actin organization via the interaction of p33/p92 and Cof1p.
Actin-organization function of Cof1p is needed for inhibition of tombusvirus replication
The best-characterized function of the multifunctional Cof1p is to bind to ADP-bound actin and depolymerize actin filaments in yeast cells [54,55]. To test if the actin-organization function of Cof1p is important for the inhibitory function of Cof1p on TBSV replication, we used a group of Cof1p mutants in the yeast-based TBSV replication assay. Interestingly, a Cof1p mutant that binds poorly to G- or F-actin (cof1-22) [66] supported TBSV replication at ~3-fold higher level than wt (S1B Fig). Also, those mutants that are defective in actin organization (such as cof1-5 ts and cof1-8 ts) supported TBSV replication at ~4-to-8-fold increased levels at the semi-permissive temperature (Fig 1A and S1A Fig). In contrast, Cof1p mutants that have wt-like organization of actin cytoskeleton [such as cof1-6, cof1-10, cof1-11, cof1-12, cof1-13 and cof1-15; [66]] did not increase TBSV replication (S1C Fig). We also tested if Cof1p mutants can interact with p33 in yeast using a split-ubiquitin assay. All the Cof1p mutants tested, including cof1-5 ts, cof1-8 ts and cof1-20, interacted with p33 (S1D Fig). Altogether, these data suggest that the ability of Cof1p to organize actin filaments in yeast is important for TBSV replication.
Mutations in ACT1 increase TBSV replication in yeast and in vitro
The above data with Cof1p mutants suggested that Cof1p likely inhibits TBSV replication by organizing or remodelling actin filaments due to its ability to promote actin depolymerization and disassembly. To test this model, we used actin mutants, which are defective in actin organization under semi-permissive temperature [67], in the yeast-based TBSV replication assay. Six yeast strains carrying actin ts mutants supported ~3-4-fold higher level of TBSV replication than wt yeast did at the semi-permissive temperature (Fig 4A–4C). These yeast mutants produced p33 and p92 to a comparable level with wt yeast (Fig 4A–4C). Overall, these actin mutants behaved similarly to selected cofilin mutants (Fig 1 and S1 Fig) by supporting high level of TBSV replication, thus suggesting that cofilin and the actin mutants might promote TBSV replication in some similar ways.
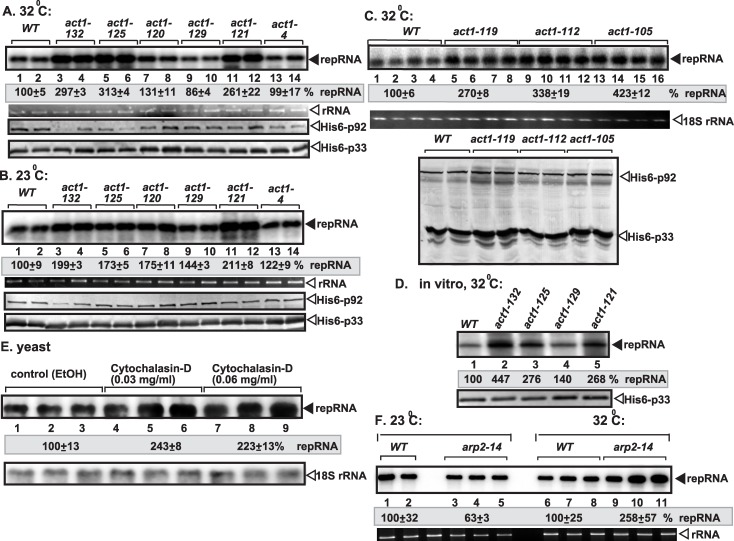
Fig 4. Act1p inhibits TBSV repRNA accumulation in yeast.
(A-C) Accumulation of TBSV repRNA in various act1 ts mutants or wt yeast at the semi-permissive (32°C, panel A), the permissive (23°C, panel B) or the semi-permissive (34°C, panel C) temperatures. To launch TBSV repRNA replication, we expressed His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter in the parental (BY4741) and act1 ts yeast strains. Northern blot analysis was used to detect DI-72(+) repRNA accumulation, which was normalized based on 18S rRNA. Each experiment was repeated three times. Bottom two panels: Western blot analysis of the accumulation level of His6-p33 and His6-p92 proteins using anti-His antibody. (D) Testing TBSV in vitro replication assay based on CFEs. The CFEs were prepared from wt or various act1 ts yeasts (cultured at the semi-permissive temperatures) expressing the tombusvirus His6-p33 and His6-p92 replication proteins. Then, the same amount (1 μg) of DI-72(+) repRNA was added to each reaction, followed by in vitro tombusvirus replication assay. The denaturing PAGE analysis shows the level of TBSV replication in the various CFEs as indicated. (E) The effect of Act1 inhibitor (Cytochalasin-D) on viral RNA accumulation in wt yeast. Northern blot analysis was used to detect DI-72(+) repRNA accumulation in yeast strain treated with Cytochalasin-D to inhibit Act1p function. Bottom panel: Northern blot analysis of the total ribosomal level in the above yeast samples. (F) Accumulation of TBSV repRNA in arp2-14 ts mutants or wt yeast at the permissive (23°C) or the semi-permissive (32°C) temperatures. See further details in panel A.
Isolation of the membrane fraction of yeasts containing the VRCs, followed by testing for viral RNA synthesis in vitro, revealed that the viral replicase from three yeast strains expressing actin ts mutants supported 2-4-fold increased TBSV repRNA replication (Fig 4D). The in vitro data are from normalized replication assays that are based on comparable level of p33 replication proteins. Altogether, the in vitro data also support that Act1p mutations have similar effects on TBSV replication to Cof1p mutants.
To further demonstrate the active role of the actin network in TBSV replication, we applied Cytochalasin-D inhibitor in yeast, which blocks the polymerization of new actin filaments [59]. We found that the treatment with the inhibitor led to ~2.5-fold increase in TBSV replication (Fig 4E). Similarly, ts mutation in Arp2, which is an actin polymerization factor [58], resulted in ~2.5-fold increase in TBSV replication at the semi-permissive temperature (Fig 4F). These data confirmed that inhibition of new actin filament polymerization needed for dynamic actin remodelling in cells stimulates TBSV replication.
Inhibition of actin organization increases TBSV replication in plant cells
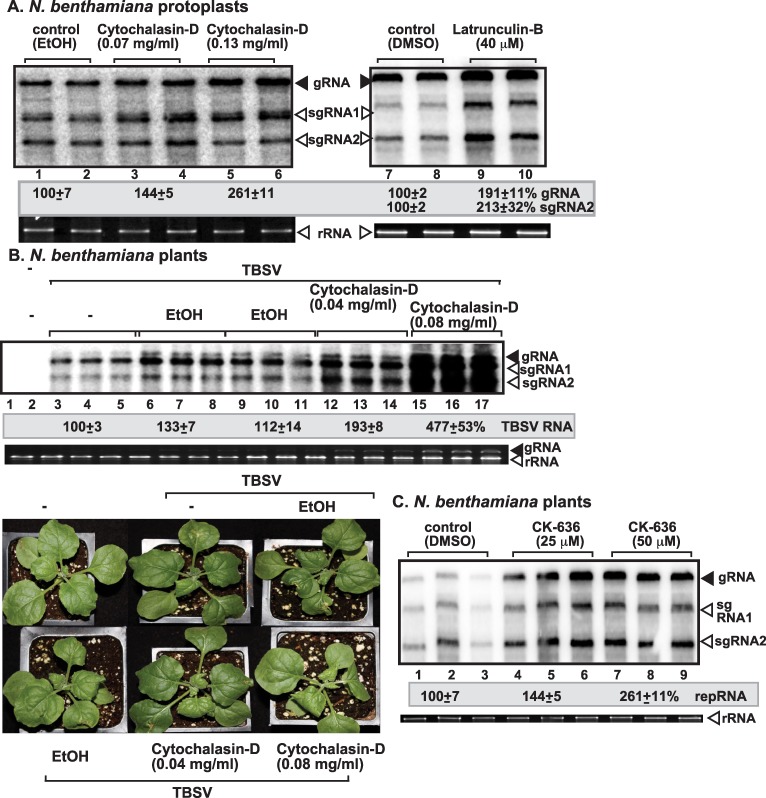
To validate whether our findings with yeast cells on the role of Cof1p and actin in TBSV replication are valid in plant cells, first we tested the effect of Cytochalasin-D and Latrunculin-B inhibitors [68] of actin organization and new actin filament formation on TBSV replication. Treatments of N. benthamiana protoplasts with these inhibitors led to ~2-to-2.5-fold increase in TBSV genomic (g)RNA accumulation (Fig 5A). These data suggest that dynamic actin network is inhibitory to TBSV replication in single plant cells.
Fig 5. The effect of actin inhibitors on TBSV RNA accumulation in N. benthamiana protoplasts and plants.
(A) Northern blot analysis was used to detect genomic (g)TBSV RNA accumulation in protoplasts treated with Cytochalasin-D (lanes 3–6) and Latrunculin-B (lanes 9–10) to inhibit dynamic actin functions. Protoplasts from N. benthamiana were electroporated with TBSV gRNA and treated with various concentrations of the inhibitors as shown. Comparable concentrations of the solvents were used as controls. Total RNA samples were obtained 24 hours post-electroporation. The ethidium-bromide stained gel at the bottom shows ribosomal RNA as loading control. The two subgenomic RNAs made during the infection process are also detected. The accumulation level of TBSV RNA was normalized based on the rRNA. Each experiment was repeated three times. (B) The effect of actin inhibitor on TBSV RNA accumulation in N. benthamiana leaves. Northern blot analysis was used to detect gTBSV RNA. Treatment of N. benthamiana leaves with Cytochalasin-D promotes the accumulation of TBSV RNAs. Total RNA samples from the inoculated leaves were obtained 3 and 4 days post inoculation and used for Northern blotting (top panel) and gel analysis to show ribosomal RNA level. Symptom intensification caused by TBSV infection in plants treated with Cytochalasin-D. Note that leaf curling is more pronounced in TBSV-infected plants 3 days post-inoculation. (C) The effect of Arp2/Arp3 inhibitor on TBSV RNA accumulation in N. benthamiana leaves. Northern blot analysis shows gRNA and sgRNAs accumulation. See further details in panel B.
Treatment of whole leaves of N. benthamiana with Cytochalasin-D also resulted in increased TBSV RNA accumulation by up to ~5-fold in the treated leaves. This was followed by the appearance of symptom intensification in the treated plant compared to the control (ethanol-treated) plants (Fig 5B). Treatment of whole leaves of N. benthamiana with CK-636, which is an inhibitor of Arp2/3 [69] also resulted in ~2.5-fold increase in TBSV genomic (g)RNA accumulation (Fig 5C). Similarly, knocking down NbAdf2 (the homolog of the yeast Cof1p) by VIGS in N. benthamiana also led to ~3-fold increase in TBSV gRNA accumulation in infected leaves (Fig 6A). This ultimately led to the rapid death of TBSV-infected N. benthamiana plants when compared with the empty vector agroinfiltrated plants (Fig 6A). Arabidopsis genome codes for four highly conserved ADF genes, which are 83-to-88% identical in nucleotide sequences [62–64], making it highly likely that our VIGS approach knocked down the expression levels of all ADF genes.
Fig 6. Knock-down of ADF2 mRNA level by VIGS increases the accumulation of TBSV RNA in N. benthamiana.
(A) Total RNA samples obtained from N. benthamiana leaves silenced as shown were analyzed by Northern blotting, which shows the accumulation of TBSV gRNA and sgRNAs. Middle image: the ethidium-bromide stained gel shows ribosomal RNA level. We chose the 10th day after VIGS to inoculate the upper, systemically-silenced leaves with TBSV virions. Samples for RNA extractions were taken 4 days post inoculation from the inoculated leaves. Bottom image: Symptoms of TBSV infected N. benthamiana plants silenced for ADF2 14 days after agroinfiltration 4 days after inoculation with TBSV virion preparation. The control experiments included the TRV1 vector as well as infiltration with water (“-”). (B) Expression of dominant negative myosin motor protein inhibits tombusvirus accumulation in N. benthamiana leaves. The C-terminal tail of myosin XI-K was expressed via agroinfiltration, followed by launching tombusvirus infection 24 h latter via agroinfiltration in the same leaves to minimize the role of cell-to-cell movement in the infiltrated leaves. Samples for Northern blotting were taken 3 days after agroinfiltration with the CNV construct. Expression of RFP via agroinfiltration in N. benthamiana leaves was used as a contol.
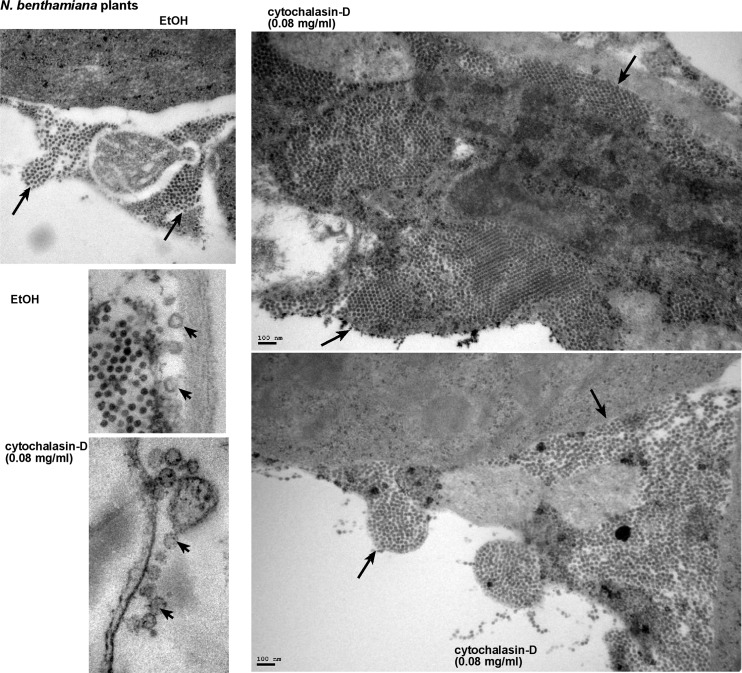
Electron microscopy (EM) images of plant cells from whole plants treated with Cytochalasin-D showed large areas of tightly packed virions in the cytosol (Fig 7). Comparable areas in the control plant cells contained fewer numbers of virions, which were packed less tightly together. However, it seems that the sizes of the virus-induced spherules (containing VRCs) were comparable in the Cytochalasin-D- treated and control plant cells (albeit based on only a small number of spherules, Fig 7). The increased number of virions in Cytochalasin-D-treated plant cells was in agreement with the higher accumulation level of TBSV RNAs in Cytochalasin-D-treated than in control plants (Fig 5). Based on all these data, the roles of cofilin (i.e. Adf2 in plants) and actin organization in replication of the TBSV genomic RNA have been firmly established in plants.
Fig 7. The presence of large number of TBSV virions in N. benthamiana cells treated with Cytochalasin-D.
Representative electron microscopic images of stained ultra-thin sections of N. benthamiana cells. N. benthamiana leaves were inoculated with TBSV, and one day later, the same leaves were infiltrated with ethanol or Cytochalasin-D at 80 μg/ml concentration using a syringe to inhibit actin polymerization. Sample preparations from the treated leaves were done two days afterwards. Several characteristic TBSV virion-containing areas are marked with arrows. Close up view of spherules, marked by arrowheads, in plant cells infected with TBSV. EtOH-treated plants were used as a control. Bars in panels represent 100 nm, while the size of the TBSV virions is 30 nm.
Tombusvirus replication sites are located in the vicinity of actin filaments in yeast and plant cells
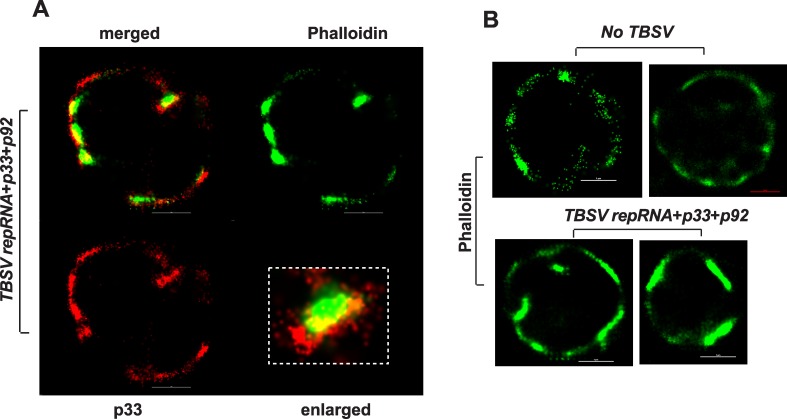
To study the putative connection between the actin network and tombusvirus replication sites, we performed super-resolution microscopy on wt yeast cells actively replicating TBSV repRNA. The replication sites were detected with anti-p33 antibody, which also recognizes p92pol because of the overlapping sequence in the pre-readthrough region in p92. The actin filaments were detected by phalloidin (F-actin specific reagent) attached to Atto-488 dye. Interestingly, several large tombusvirus replication sites were located in the vicinity of the actin filaments in wt yeast (Fig 8A and S1 Video). Also, the actin filaments formed larger structures (called “patches” with up to 2-fold increase in diameter) in yeast cells replicating TBSV than in the absence of viral components (Fig 8B).
Fig 8. Super Resolution laser microscopic images of yeast cells.
(A) A yeast cell replicating TBSV repRNA was imaged based on Alexa Flour 647 (for p33 replication protein) and for ATTO488-phalloidin, which detect actin filaments. The bars represent 1 μm. The boxed area represents enlargement of the image to visualize the localization of actin and p33 replication protein in yeast. (B) Yeast cells free of viral components (top images) or replicating TBSV repRNA (bottom images) were imaged based on ATTO488-phalloidin, which detect actin filaments. The bars represent 1 μm. The images in both panels were prepared by a Nikon Super Resolution Microscope N-STORM and image processing was performed using NIS-element software.
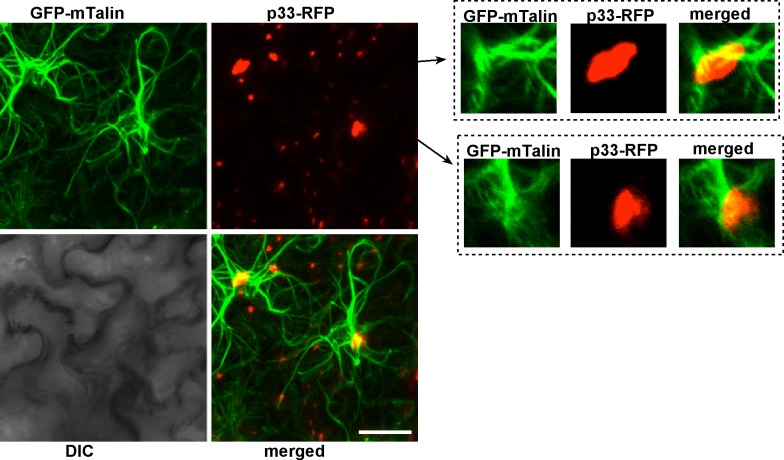
Because the actin cables are difficult to visualize in the tiny yeast cells, we also used plant cells transiently expressing RFP-tagged p33 replication protein. The actin cables were visualized via transgenically expressed GFP-tagged mTalin, which binds to actin filaments [70]. Confocal laser microscopy imaging revealed that the largest TBSV replication sites (marked by p33-RFP), which are known to form on aggregated peroxisomal membrane surfaces, were formed where actin cables intersected (Fig 9). Individual cross-sections showed that the actin cables surrounded and sometimes crossed through the large viral replication sites/organelles (Fig 9, images on the right, and S3 Fig, which shows the structures in the presence of TBSV infection). Interestingly, we observed very similar arrangements between actin cables and viral replication sites (marked by p36-RFP) in case of Carnation Italian ringspot virus (CIRV), a closely related tombusvirus that replicates on mitochondrial membrane surfaces (Fig 10 and S4 Fig, which shows the structures in the presence of CIRV infection). Comparison of actin filament network revealed overall higher density and thicker and larger number of individual filaments in TBSV-replicating (S3 Fig versus S5 Fig), and to a lesser extent, CIRV-replicating plant cells (S4 Fig) than in uninfected control cells (S5 Fig). Thus, the actin network plays a role in formation of tombusvirus replication sites that resemble replication organelles in plant cells [71].
Fig 9. Confocal laser microscopic images of plant cells expressing TBSV p33 replication protein.
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing p33-RFP. The images on the left represent Z-stack images (overlay of individual images), while on the right, represent single plane images. Note the localization of large p33-RFP containing areas (i.e., replication organelles) at the intersection of actin cables. The bar represents 20 μm.
Fig 10. Confocal laser microscopic images of plant cells expressing CIRV p36 replication protein.
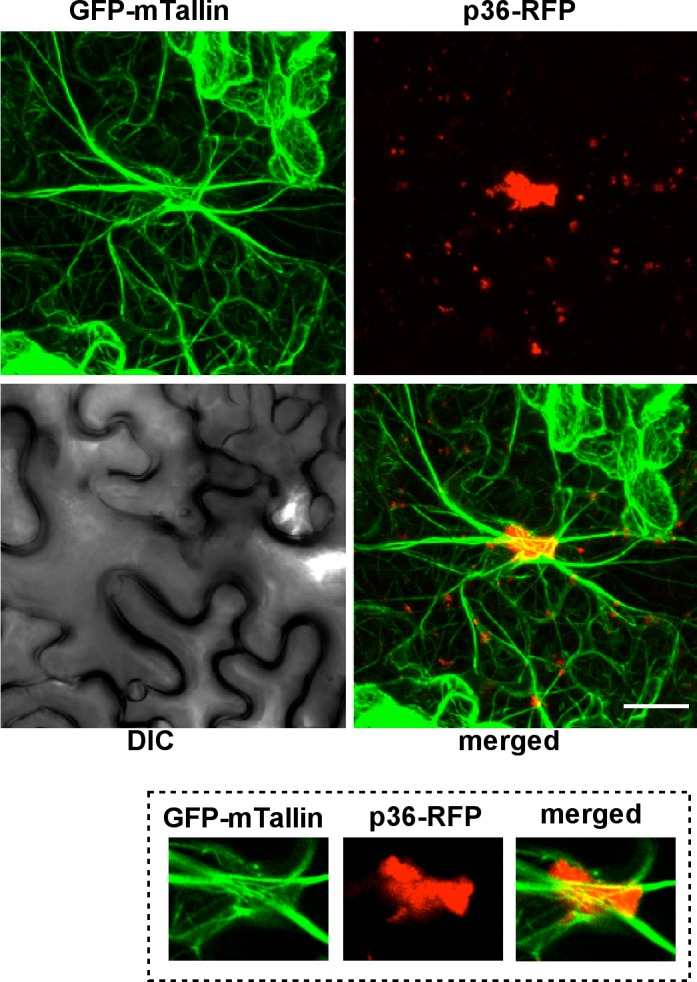
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing the CIRV p36-RFP. The images on the top represent Z-stack images, while on the bottom, represent single plane images. Note the localization of large p36-RFP containing areas (i.e., replication organelles including mitochondria) at the intersection of actin cables. The bar represents 20 μm.
The close association between the actin network and tombusvirus replication proteins in both yeast and plant cells indicated a putative functional role for actin in tombusvirus replication. Since both cofilin and actin are essential for yeast and plant cells, in the above experiments we used approaches that temporarily and partially blocked the dynamic actin network, but still allowed the function of the stable actin network. To test the putative role of the actin network, we inhibited the functions of both dynamic and stable actin networks via expressing dominant negative mutant of myosin XI-K that is a motor protein involved in actin-based moving of cargos in the cytosol of plant cells [72,73]. Interestingly, expression of a dominant negative mutant of myosin XI-K strongly inhibited tombusvirus replication in N. benthamiana leaves (Fig 6B). All these data support the functional role of the actin network in tombusvirus replication.
Tombusvirus replication takes advantage of altered sterol transport in yeast with mutant cofilin or actin
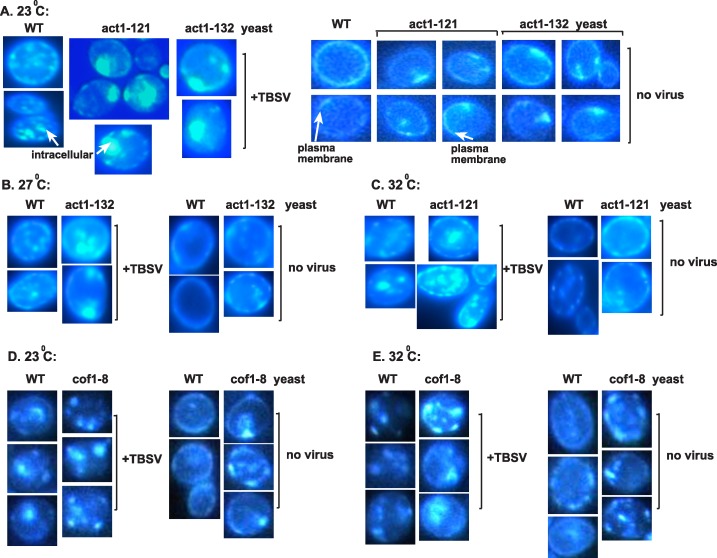
Because TBSV replication greatly depends on the virus-driven retargeting of sterols, which become highly enriched at viral replication sites and facilitate the efficient assembly of VRCs [45,47], we analyzed the sterol distribution in cofilin and actin mutant yeasts. Filipin-based staining of sterols, which can be visualized by fluorescent microscopy [45], revealed the formation of large sterol-enriched compartments in cofilin and actin mutant yeasts when compared to wt yeast showing smaller sterol-enriched compartments (Fig 11). The difference in sterol enrichment and distribution was especially striking in the presence of viral components in cof1-8 ts (Fig 11E), act1-132 ts and act1-121 ts (Fig 11B and 11C) yeasts in comparison with the wt yeast at the semi-permissive temperature. We have shown previously that these internal (not plasma membrane localized as observed in the absence of viral components in wt yeast, Fig 11A) sterol-enriched compartments represent the viral replication sites [45].
Fig 11. The cofilin and actin mutants facilitate the enrichment of sterols at the sites of tombusvirus replication in yeast.
(A) Re-localization of ergosterols to internal punctate structures in yeast replicating TBSV repRNA. Fluorescent microscopic images of yeast cells stained with filipin dye. Note that filipin stains ergosterols present mostly at the plasma membrane in virus-free wt yeast cells, while act1-121 ts yeast and act1-132 ts yeast show uneven distribution of ergosterols in the absence of viral components (right images). (B) Similar experiments as in Panel A, except performed at the semi-permissive temperature (27°C) in act1-132 ts and WT yeasts. (C) Similar experiments as in Panel A, except performed at the semi-permissive temperature (32°C) in act1-121 ts and WT yeasts. (D) Re-localization of ergosterols to internal punctate structures in yeast replicating TBSV repRNA in cof1-8 ts or wt yeasts at the permissive temperature. Fluorescent microscopic images of yeast cells stained with filipin dye. (E) Re-localization of ergosterols to internal punctate structures in yeast replicating TBSV repRNA in cof1-8 ts or wt yeasts at the semi-permissive temperature. See further details in panel A. Each experiment was repeated at least three times.
In comparison with the wt yeast, the cofilin and actin mutant yeasts showed less even distribution of sterols in the plasma membrane, suggesting defect in normal sterol transport in case of the cofilin and actin mutants (Fig 11). Based on these observations, we suggest that TBSV could take advantage of the defective sterol transport mechanism in cofilin and actin mutant yeasts that likely allows the virus to easily hijack sterols and enrich them at the sites of viral replication that leads to highly efficient VRC assembly and increased viral RNA synthesis.
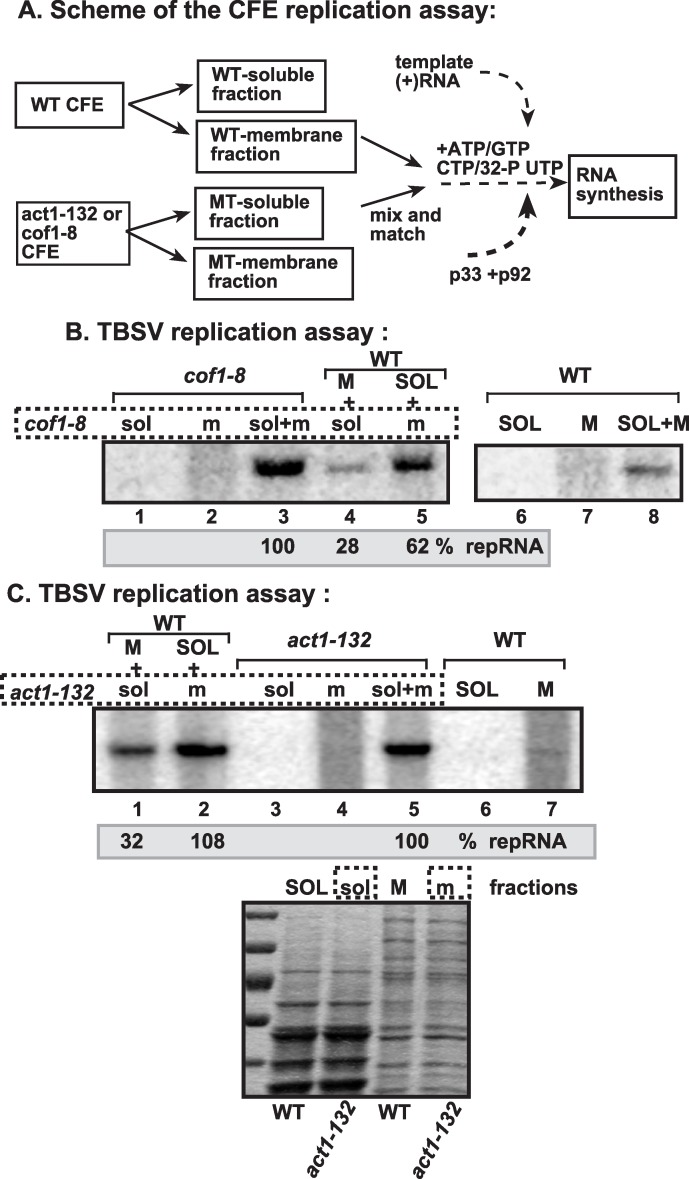
To examine if the membrane fraction from the cof1-8 ts or act1-132 ts yeasts is more suitable for TBSV replication than from wt yeast at the semi-permissive temperature, we used a mix-and-match CFE approach as shown schematically in Fig 12A. The combination of membrane fraction from cof1-8 ts with the soluble fraction from the wt yeast (Fig 12B, lane 5) supported TBSV repRNA replication almost as efficiently as the nonfractionated CFE from cof1-8 ts (Fig 12B, lane 3). On the contrary, the combination of membrane fraction from wt yeast with the soluble fraction from cof1-8 ts supported TBSV repRNA replication at a ~4-fold reduced level (Fig 12B, lane 4), suggesting that the membrane fraction of cof1-8 ts is responsible for the enhanced TBSV replication in vitro. We obtained similar results with the membrane fraction of act1-132 ts (Fig 12C, lane 5), which was responsible for the enhanced level of TBSV repRNA replication in vitro (Fig 12C). Altogether, these in vitro data are in agreement with the model that the intracellular membranes in cof1-8 ts or act1-132 ts yeasts are more suitable for TBSV replication, possibly because they are sterol-rich, in comparison with the intracellular membranes in wt yeast.
Fig 12. The membrane fraction of cofilin and actin mutant yeasts is critical for tombusvirus replication in vitro.
(A) The scheme of the CFE-based TBSV replication assay. The membrane and soluble fractions of the CFEs prepared from wt, cof1-8 ts or act1-132 ts yeasts were used alone or in combinations (as shown). Purified recombinant MBP-p33 (7 pmol) and MBP-p92pol (4 pmol) replication proteins in combination with DI-72 (+)repRNA (0.5 μg) were added to the CFEs. (B) Denaturing PAGE analysis of the 32P-labeled TBSV repRNA products obtained is shown. Note that, prior to the TBSV replication assay, the membrane and soluble fractions of CFEs derived from wt or cof1-8 ts yeasts were adjusted to contain comparable amounts of the cellular proteins. Note that the enhanced TBSV repRNA replication in CFE prepared from cof1-8 ts yeast depends on the membrane fraction. (C) Comparable CFE-based TBSV replication studies as in panel B, except using CFE prepared from act1-132 ts yeast. Bottom image shows coomassie-stainde SDS PAGE of the total proteins in the membrane and soluble fractions of CFEs. Note that the enhanced TBSV repRNA replication in CFE prepared from act1-132 ts yeast depends on the membrane fraction.
Efficient recruitment of oxysterol binding protein and VAP protein by TBSV in yeast with mutant cofilin or actin
TBSV controls intracellular sterol transport via hijacking oxysterol binding proteins (OSBP-like or ORP, such as Osh3p, Osh5p, Osh6p and Osh7p) and VAP protein (VAMP-associated protein, such as yeast Scs2p or Vap27-1, a plant ortholog) via interaction with the p33 replication protein [45]. These cellular proteins help TBSV to stabilize membrane contact sites (MCSs) between the ER and the peroxisome, thus facilitating the efficient transport of sterols from the ER to the peroxisomal membranes where a large number of VRCs forms [45].
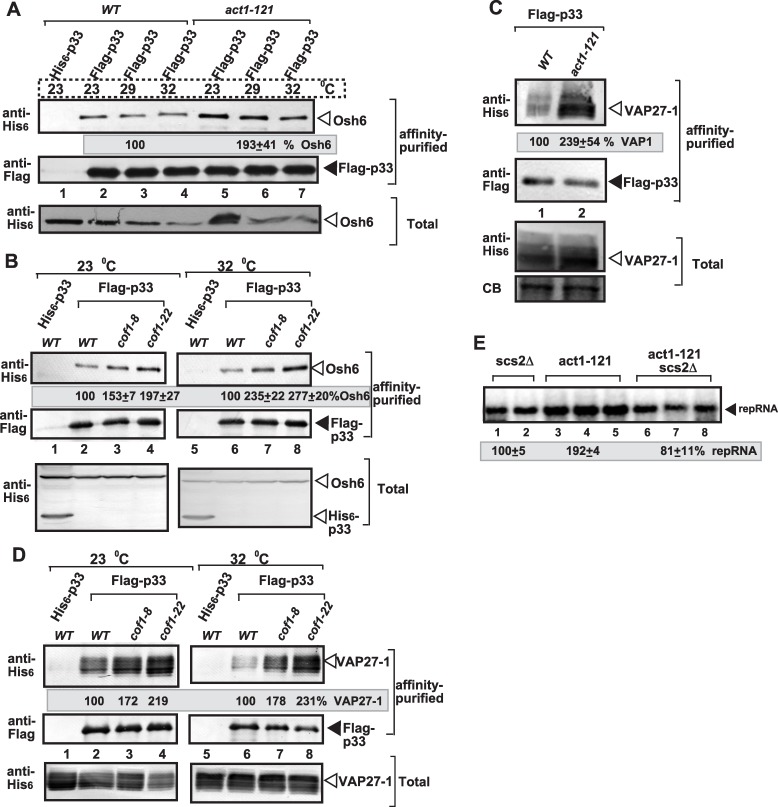
To test if TBSV could efficiently co-opt Osh6p and Vap27-1 [45] in yeast cofilin or actin mutants, we purified p33 replication proteins from detergent-solubilized membrane fractions, followed by Western blotting to measure the co-purified cellular Osh6p and Vap27-1. We found ~2-to-2.5-fold increase in the co-purified cellular Osh6p in cof1-8 ts, cof1-22 ts (Fig 13B), act1-121 ts (Fig 13A) and act1-132 ts (Table 1) in comparison with wt yeast at the semi-permissive temperature. The Osh6p protein was also more efficiently co-purified with p33 in act1-105 ts, and act1-112 ts yeasts in comparison with wt yeast at the permissive or semi-permissive temperature (S6 Fig). Interestingly, we also found ~2-to-2.5-fold increase in the co-purified cellular Vap27-1, in cof1-8 ts, cof1-22 ts (Fig 13D), act1-121 ts (Fig 13C) and act1-132 ts (Table 1) in comparison with wt yeast at the semi-permissive temperature. Altogether, the more efficient co-purification of Osh6p and Vap27-1 in the cofilin and actin mutants suggests the enhanced or more stable formation of viral-induced MCSs and could explain the high enrichment of sterols at internal sites (likely representing the replication sites) in the cofilin and actin mutants.
Fig 13. Cof1p and Act1p mutations affect the subversion of MCS forming OSBP-like Osh and VAP proteins for tombusvirus replication in yeast.
(A) Enhanced co-purification of yeast Osh6p ORP with the p33 replication protein from act1-121 ts yeast in comparison with WT yeast. The FLAG-tagged p33 and FLAG-p92 were co-purified from solubilized membranous fraction of yeast extracts using a FLAG-affinity column. Yeast expressing the His6-tagged p33 was used as a control. Top panel: Western blot analysis of the co-purified 6xHis-tagged Osh6p with anti-His antibody in the affinity-purified preparations. Middle panel: Western-blot analysis of the same samples as in the top panel, but using anti-FLAG antibody. Bottom panels: Western blot analysis of 6xHis-Osh6p with anti-His antibody in the total protein extract from yeast expressing the shown proteins. (B) Enhanced co-purification of yeast Osh6p ORP with the p33 replication protein from cof1-8 ts yeast in comparison with WT yeast at the permissive and semi-permissive temperatures. See further details in panel A. (C) Enhanced co-purification of the ER-localized Vap27-1 with the p33 replication protein from act1-121 ts yeast in comparison with WT yeast at the semi-permissive temperature. The FLAG-affinity purification was done as in panel A. Top panel: Western blot analysis of the co-purified 6xHis-Vap27-1 with anti-His antibody in the affinity-purified preparations. Middle panel: Western-blot analysis of the same samples as in the top panel, but using anti-FLAG antibody. Bottom panels: Western blot analysis of 6xHis-Vap27-1 with anti-His antibody in the total protein extract from yeast expressing the shown proteins. CB: Coomassie-stained SDS-PAGE of total protein extract. Each experiment was repeated two times. (D) Enhanced co-purification of the ER-localized Vap27-1 with the p33 replication protein from cof1-8 ts yeast in comparison with WT yeast at the permissive and semi-permissive temperatures. See further details in panel A. (E) Dependence of TBSV repRNA replication on Scs2p VAP protein in yeast. The double-mutant yeast carrying act1-121 ts mutation and missing SCS2 shows reduced replication of TBSV repRNA based on Northern blot analysis. See further details in Fig 1.
Table 1. Co-purified host proteins with p33 from act1-132 yeast.
| Protein | percentage |
|---|---|
| Ded1 helicase | 108±9 |
| Cdc34 E2 enzyme | 207±22 |
| Osh6 ORP | 258±37 |
| Pbp2 RNA binding | 85±18 |
| Rpn11 deubiquitinase | 161±12 |
| Tdh2 GAPDH | 111±15 |
| Tef1 eEF1A | 96±8 |
| Vap27-1 (Scs2-like) | 218±33 |
| Vps4 AAA ATPase | 53±6 |
| Vps23 ESCRT I | 108±9 |
Co-purification of the given host protein with p33 from WT yeast is taken as 100%.
To further test if indeed the actin mutants affect sterol-enrichment at viral replication sites, we deleted Scs2p VAP protein in yeast (double mutant act1-121 ts scs2Δ yeast), followed by measuring TBSV repRNA replication. Interestingly, the double mutant supported a reduced level of repRNA replication, comparable to that observed in the single mutant yeast (scs2Δ) (Fig 13E). These data suggest that the actin mutant could enhance TBSV replication only when functional VAP protein is present, thus when MCSs are formed, in act1-121 ts yeast.
The co-purification-based proteomics approach also revealed that co-opted host factors, such as Cdc34 ubiquitin-conjugating enzyme, Rpn11 deubiqutinase and Vps23p ESCRT factor, were co-purified with the tombusvirus replicase by ~2-fold more efficiently from act1-132 ts yeast in comparison with wt yeast (Table 1). Other cellular host factors, such as Tef1p (eEF1A), Tdh2p (GAPDH), DDX3-like Ded1p and Vps4p AAA ATPase, were co-purified with p33 as efficiently from act1-132 ts yeast as from wt yeast (Table 1). Based on these data, we suggest that the actin mutations also facilitate the subversion of some cellular host factors by TBSV into the VRCs.
Discussion
Tombusvirus replication involves the assembly of hundreds-to-thousands of VRCs in infected cells that leads to robust viral RNA synthesis [45,71]. The VRCs are clustered in large viral-induced compartments, called replication organelles [74]. The formation of individual VRCs, housed in spherule-like membranous structures, requires p33 and p92 replication proteins, the viral (+)RNA and 15–20 co-opted host proteins, lipids, such as sterols and phospholipids, and likely metabolites [3,16,43]. TBSV replication also depends on MCSs to transfer sterols to the sites of replication, which is the peroxisomal membrane for TBSV and mitochondrial outer membrane for CIRV [45]. How can all these viral and cellular components come together at the sites of replication and facilitate the formation of large replication organelles containing relatively stable membranous VRCs inside the cytoplasm? Based on the current work, it seems that the actin network plays a key role in this critical viral process. Accordingly, the large p33 containing replication structures are located in the close vicinity of actin patches in yeast cells or around actin cable hubs in infected plant cells (Figs 8–10). Therefore, these actin filaments are likely involved in VRC assembly and the formation of large viral replication organelles containing many individual VRCs. Indeed, expression of dominant negative mutant of myosin XI-K motor protein, which is involved in actin-based intracellular cargo movement [72,73], strongly inhibited tombusvirus accumulation in N. benthamiana leaves, supporting the functional role the actin network in tombusvirus replication (Fig 6B).
Yet, several mutations in the essential ACT1 gene or the use of actin inhibitors enhance TBSV replication in yeast, suggesting that ACT1 is an antiviral restriction factor. However, the identified mutations and the inhibitors affect the assembly of new actin filaments without blocking the functions of the existing actin filaments [67,75]. Therefore, we propose that TBSV do utilize the existing actin filaments to concentrate host and viral components at replication sites and assemble VRCs. Accordingly, the large tombusvirus replication organelles are frequently formed where the actin cables meet in the cytosol for both peroxisomal TBSV and the mitochondrial CIRV in plant cells (Figs 9 and 10 and S3–S5 Figs). On the other hand, the dynamic rearrangement of the actin filaments might limit the VRC assembly process or used in antiviral responses by the cell, making new actin filament formation inhibitory to TBSV replication. Altogether, the emerging picture from all our data is that the dynamic actin network functions as a virus restriction factor by inhibiting tombusvirus replication, while the stable actin network provides pro-viral function in tombusvirus VRC formation. This could be a more general phenomenon, because disruption of the actin network leads to enhanced susceptibility to pathogenic bacteria due to inhibition of the innate immunity response of the plant [76].
Interaction between the cellular cofilin and viral replication factor regulates actin organization and promotes TBSV replication
Using tombusviruses, we have discovered that the cellular cofilin actin depolymization factor is targeted by TBSV to obstruct the dynamic actin network. Down-regulation of Cof1p level or the use of Cof1p ts mutants at the semi-permissive temperature lead to increased level of TBSV RNA accumulation in yeast cells. The increased RNA replication is due to highly active VRCs, based on testing the in vitro activity of replicase in CFE prepared from yeast expressing cof1-8 ts, which was ~3-fold more active than the replicase operating in wt CFE (Fig 1D). Also, expression of NtAdf2, a plant homolog of Cof1p [62,63] reduced TBSV replication in vitro (Fig 2D).
It seems that the inhibitory effect of Cof1p on TBSV replication is due to two different mechanisms. First, the interaction between Cof1p (Adf2 homolog) and p33 and possibly p92 could partially sequester the replication proteins, thus limiting their participation in VRC formation and TBSV RNA replication. Second, Cof1p also seems to inhibit TBSV replication via facilitating the dynamic actin re-organization in live yeast cells. The Cof1p-driven disassembly of existing actin filaments in combination with formation of new actin filaments might inhibit the continuous growth of the replication compartments, which could depend on stable MCSs for sterol transfer/enrichment at the sites of replication and steady supply of host factors to the replication sites (Fig 14). Accordingly, yeasts expressing Act1p ts mutants at the semi-permissive temperature supported increased level of TBSV RNA accumulation (Fig 4). Moreover, pharmacological inhibition of actin organization by Cytochalasin D or Latrunculin B in yeast, plant protoplasts or whole plants also resulted in increased level of TBSV RNA accumulation (Figs 4–7). Thus, actin organization, especially the formation of new actin filaments is a major restriction factor for virus replication and TBSV modulates the dynamic actin network to obstruct the formation of new actin polymers.
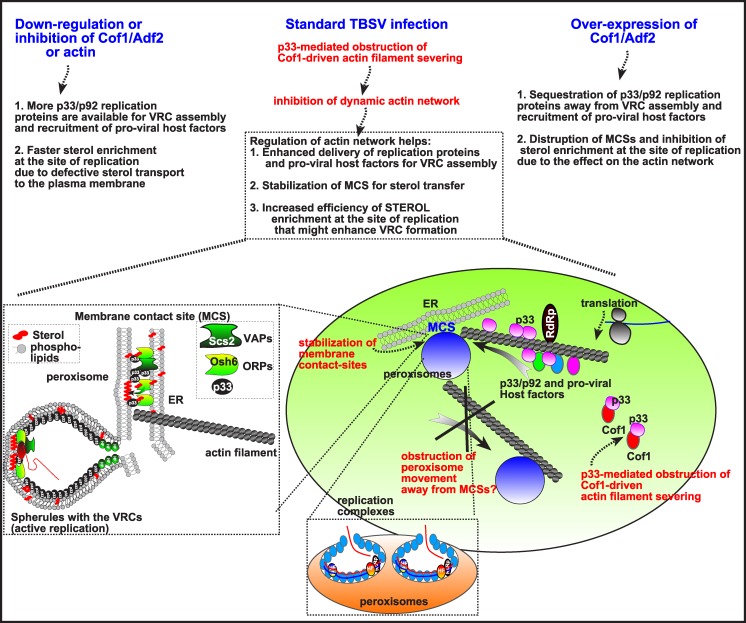
Fig 14. Summary and model on the role of p33 replication protein driven obstruction of the cellular actin network in TBSV replication.
Top: three scenarios with different levels of Cof1p are shown, including the predicted effects on TBSV replication. Bottom: The formation of VRCs (virus-induced membrane invaginations, called spherules) in the peroxisomes requires the subversion of cellular co-factors and lipids, whose recruitment could be enhanced by obstructing new actin filament formation via p33:Cof1p interaction. In addition, formation and/or stabilization of membrane contact sites (MCS) between ER and peroxisome might be promoted by blocking the dynamic actin network, and halting the movement of peroxisomes away from ER to various subcellular locations. Moreover, obstructing the dynamic actin network might also facilitate localized enrichment of sterols within VRCs, which is known to be required for tombusvirus VRC formation. Left: Enlargement of the scheme showing the predicted MCS with co-opted cellular ORP and VAP proteins that facilitate tombusvirus replicase assembly (shown as a vesicle-like structure) [45]. The OSBP-like ORP oxysterol-binding proteins are recruited via binding to the tombusviral p33 replication protein to the ER-bound Scs2p VAP to form MCS between the ER and peroxisomal membranes. The ORPs then facilitate the enrichment of sterols in the peroxisomal membrane, forming sterol-rich microdomains needed for the formation of virus-induced spherules. These spherules contain the VRCs performing viral RNA synthesis.
How does dynamic actin organization limit TBSV replication in live cells? Based on our current knowledge on TBSV replication, 15–20 cytosolic pro-viral host factors, peroxisomes/ER, phospholipids and sterols are required for efficient TBSV replication [9,16,17,46,47,77–79]. We predict that TBSV could easily co-opt these host factors for efficient VRC assembly when dynamic actin organization and remodeling of the actin network is inhibited. The obstructed actin network might inhibit the normal cellular distribution and function of host proteins and organelles, which might become more readily accessible to TBSV when new actin filament formation is hindered (Fig 14). Indeed, formation of large replication foci in combination with increased replicase activity is seen in cof1-8 ts yeast at the semi-permissive temperature (Fig 3). Similar observation of the presence of large replication foci and increased replication was documented for yeast over-expressing Ino2p, a transcription factor regulating phospholipid biosynthesis [46]. Therefore, our model predicts that it is more difficult for the virus to recruit the host factors, sterols and peroxisomes for efficient VRC assembly when dynamic actin organization facilitates rapid transport of host components to their final destination within the cell, while disruption of actin organization via p33-Cof1p interaction and Cof1p re-localization (in natural infections) or via inhibitors, actin or Cof1p mutations enhances VRC assembly and TBSV replication (Fig 14). The actin network might also regulate the availability of given host factors, such as eEF1A, whose function is affected by actin through promoting GTP hydrolysis by the GTP-bound eEF1A and subsequent release of eEF1A from translation [80] that might increase the availability of eEF1A for viral functions [38,81].
Cofilin not only regulates actin dynamics, but also affects phospholipid metabolism via regulating phospholipase D activity [53,54,56,57]. However, previous genomics and proteomics screens with TBSV have not identified SPO14 or other phospholipase genes [9,26–30,32,34]. Therefore, the evidence is lacking for this mechanism during TBSV replication. Also, p33 interaction with Cof1p might interfere with apoptosis that requires re-localization of cofilin to the mitochondria [54].
Since other (+)RNA viruses also remodel subcellular membranes and rewire cellular pathways to facilitate virus replication and avoid antiviral responses [74], it is possible that targeting cofilin and actin could be wide-spread among viruses. Accordingly, during HIV infection, the viral Nef protein inactivates cofilin molecules via its interaction with Pak2 cellular kinase that phosphorylates cofilin [82]. This results in reduced fibroblast and T cell motility, thus helping HIV to overcome T lymphocytes-mediated, chemotaxis-based antiviral response of the host. Also, HIV activates cofilin through chemokine receptor signaling to mediate entry into resting CD4 T cells [83]. These examples together with TBSV-mediated inhibition of cofilin/actin functions suggest that the actin network could be a major target for viruses to facilitate their replication in host cells. Cofilin and actin are also major targets of bacterial pathogens infecting mammalian cells, suggesting that the dynamics of actin network is key component and pathogenicity determinant [84].
Summary
Virus-induced blocking of actin organization facilitates efficient VRC assembly
We show that tombusviruses actively interfere with actin organization during replication. The mechanism of interference is based on interaction of the viral p33 and possibly p92 replication proteins with Cof1p. Since Cof1p is a key actin depolymerization factor [54,55], binding of p33 to Cof1p could inhibit Cof1p interaction with ADP-actin and block actin filament disassembly and recycling of actin monomers to form new actin filaments. Accordingly, co-expression of p33 and p92 in yeast led to large actin-containing areas in the cell, visible under confocal microscope, which is indicative of disorganization of actin patches and lack of dynamic actin function. Based on these observations, we propose that TBSV actively blocks actin organization and remodeling of the actin network via p33-Cof1p interaction and possibly by re-localization of Cof1p from the cytosol to membranes. This block in actin organization is likely temporal, occurring before and during the assembly of the VRC. After VRC assembly, which results in sequestration of newly made p33/p92 molecules into the VRCs, the role of Cof1p in actin organization is likely restored during regular TBSV infections since the actin is required for cell-to-cell movement of plant viruses [85,86].
Materials and Methods
Yeast strains and expression plasmids
Expression plasmids, S. cerevisiae strains, culturing conditions and TBSV repRNA measurements are presented in S1 Materials and Methods.
Treatment of yeast with Cytochalasin-D
Cytochalasin-D (Santa Cruz Biotechnology) [59] was added to overnight culture of BY4741 (transformed with pHISGBK-CUP1-p33:ADH-DI-72 and pESC-CUP1-His-p92) at a final concentration of 0.03 and 0.06 μg/μl. The yeast was grown for further 24 h at 23°C and total RNA was extracted for Northern blot analysis.
Treatments of plants and protoplasts with Actin inhibitors
N. benthamiana leaves were first sap-inoculated with TBSV, and one day later, the same leaves were infiltrated with ethanol or Cytochalasin-D at 80 μg/ml concentration (or as specified in Fig legends) using a syringe to inhibit actin polymerization. Two days later, total RNA was isolated from the inoculated leaves and Northern hybridization was performed to measure tombusvirus RNA accumulation.
In case of inhibitor treatment of the Arp2/3 complex, N. benthamiana leaves were infiltrated with DMSO (50 μM) or CK-636 (25 or 50 μM) (ApexBio), and 12 h later, the same leaves were inoculated with TBSV-containing sap. One day later the infected leaves were again infiltrated with DMSO (50 μM) or CK-636 (25 or 50 μM). One day later total RNA was isolated and Northern hybridization was performed as described above.
Protoplasts were isolated from N. benthamiana leaves as previously described [87]. Freshly prepared protoplasts were treated with 70 or 130 μg/ml Cytochalasin-D or 40 μM of Latrunculin-B before electroporation with 1 μg of TBSV RNA. Protoplasts were incubated in the dark for 24 h at room temperature and the total RNA was extracted for Northern hybridization [87].
Confocal laser microscopic analysis of plant F-actin in the presence of viral proteins
Expression plasmids pGD-T33-RFP or pGD-C36-RFP were transformed into Agrobacterium C58C1 separately, and transformed cells were selected on LB plate containing 50 μg/ml kanamycin, 100 μg/ml rifampicin and 5 μg/ml tetracycline. Transformed cells were grown in LB medium containing the antibiotics described above overnight, and suspended in MMA solution (10mM MES pH5.6, 10mM MgCl2, 200 μM acetosyringone) at OD600 of 1.0 for 3–4 hours. Leaves of 8 weeks old transgenic Nicotiana benthamiana plants expressing GFP-mTalin (mouse Talin) [88], which specifically binds to F-actin [70] (a gift from Dr. Michael M. Goodin at University of Kentucky), were infiltrated with Agrobacterium in MMA solution. Leaf epidermal cells were observed under confocal laser microscope (Olympus FV1000 microscope) 2 days after infiltration for localization of both GFP-mTalin and p33-RFP/p36-RFP [89]. Note that GFP-mTalin transgenic Nicotiana benthamiana plants have normal growth when compared to wild type plants under green house conditions [88]. GFP-mTalin binds to F-actin, and its localization overlaps with phalloidin stained actin filaments [70].
Super-resolution laser microscopic analysis of yeast actin in the presence of viral proteins
Yeast strain BY4741 was transformed with pESC-T33/DI72 and pYES-T92 [89]. Transformed cells were pre-grown in synthetic complete medium lacking appropriate amino acids and containing 2% glucose overnight at 29°C. Then, yeast cells were grown in synthetic complete medium lacking appropriate amino acids containing 2% galactose at a starting 0.3 OD600 for 12–14 hours at 23°C. Cells were harvested, fixed with 3.7% formaldehyde and digested with zymolase 20T to remove cell wall as described in [43]. Treated cells were applied to poly-L-lysine–coated cover slips. The cells on the cover slips were sequentially immersed in methanol for 6 min and acetone for 30s at –20°C. Anti-p33 primary antibodies (a gift from Dr. Herman B. Scholthof, Texas A&M University) were diluted (1:400) in PBS (pH 7.4) containing 0.05% Nonidet P-40 and 1% BSA and incubated overnight with the fixed cells at +4°C. Yeast cells were washed three times with PBS (pH 7.4) with 1% BSA, then incubated subsequently with anti-mouse secondary antibody conjugated to Alexa Flour 647 for 1 h and ATTO488-phalloidin for 1 h in PBS (pH 7.4) with 1% BSA, and washed three times with PBS (pH 7.4) containing 1% BSA for at least 1 h to reduce background. Cells on the cover slip were subjected to super-resolution microscopic observation (N-STORM Super Resolution Microscopy System from Nikon).
Filipin-based staining of ergoterols in yeasts
To examine the distribution of ergoterol in BY4741 and act1ts mutants (act1-121 and act1-132), the yeast strains were co-transformed with plasmids pGBK-Hisp33-CUP1/Gal DI-72 and pGAD-Hisp92-CUP1. Control untransformed and transformed yeasts were grown in SC minimal media supplemented with 2% galactose and 50 μM CuSO4 for 24 hours at 23°C, 27°C and 32°C. Cultures were fixed with 3% formaldehyde for 1 h at room temperature. Fixed cells were centrifuged and washed twice with distilled water. Washed cells were incubated with 5 mg/ml filipin complex (Sigma Chemicals) in the dark for 15 min at 23°. Filipin-based fluorescence was observed by spotting 3 μl of the cell suspensions onto poly-lysine microscope slides under UV light microscope using DAPI filter [45].
Co-purification of Osh6 protein with the p33 replication protein from yeasts
For co-purification of Osh6p protein with the membrane-bound p33 and p92 replication proteins [45], yeast strains BY4741, cof1-8 ts, cof1-22 ts, and the act1-121 ts mutants were co-transformed with plasmids HpGBK-CUP1-FLAGp33/GAL1-DI-72 and LpGAD-CUP1-FLAGp92 and (or HpGBK-CUP1-Hisp33/Gal1-DI-72 and LpGAD-CUP1-Hisp92 as a control) plus the UpYC-NT plasmids expressing His6-tagged Osh6 protein from the GAL1 promoter (UpYC-NT-Gal1-OSH6). Transformed yeasts cultures were pre-grown in selective SC-ULH− + BCS medium supplemented with 2% glucose for 12 h at 23°C and then transferred to selective medium supplemented with 2% galactose for 24 h at either 23 or 32°C to induce His6-Osh6 protein expression from the GAL1 promoter. Then the cultures were supplemented with 50 μM CuSO4 to induce expression of FLAG-p33 and FLAG-p92 or His6-p33 and His6-p92 from the CUP1 promoter, and yeast cultures were grown for an additional 4 h at 23°C or 32°C. The cultures were centrifuged washed once with phosphate-buffered saline (PBS), and then incubated in PBS buffer containing 1% formaldehyde for 1 h on ice to cross-link proteins [45]. Finally, yeast cultures were washed in PBS and proteins were FLAG-affinity purified as described previously [45].
Supporting Information
(DOC)
(A) Accumulation of TBSV repRNA in cof1-5 ts or wt yeast at the semi-permissive (32°C) temperature. To launch TBSV repRNA replication, we expressed His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter in the parental (BY4741) and cof1-5 ts yeast strains. Northern blot analysis was used to detect DI-72(+) repRNA accumulation, which was normalized based on 5S rRNA. Each experiment was repeated three times. (B) Testing Cof1p mutants defective in Act1p binding for their ability to inhibit TBSV repRNA accumulation in yeast. (C) Testing Cof1p mutants that bind Act1p for their ability to inhibit TBSV repRNA accumulation in yeast. See further details in panel A. (D) Cof1p mutant proteins interact with the TBSV p33 replication protein in yeast. The split ubiquitin assay was used to test binding between p33 and Cof1p in yeast. The bait p33 was co-expressed with the shown prey proteins. SSA1 and the empty prey vector (NubG-X) were used as positive and negative controls, respectively. The experiment with wt Cof1p is encircled.
(EPS)
(A-B) The wt or cof1-8 ts yeast co-expressed the His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter. Over-expression of the untagged native Cof1p was done from the GAL1 promoter. Top images: Northern blot analysis of TBSV repRNA in yeast samples over-expressing Cof1p or without over-expression is shown. repRNA replication took place for 24 hours at 23°C or 32°C in wt or cof1-8 ts yeast before RNA analysis. The accumulation level of DI-72(+) repRNA (shown in percentage) was normalized based on 18S rRNA.
(EPS)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing p33-RFP and infected with TBSV using sap inoculation. Note the localization of large p33-RFP containing areas (i.e., replication organelles) frequently at the intersection of actin cables. The bars represent 20 μm.
(TIF)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing p36-RFP and infected with CIRV using sap inoculation. Note the localization of large p36-RFP containing areas frequently at the intersection of actin cables. The bars represent 20 μm.
(TIF)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with an empty pGD plasmid. The bars represent 20 μm.
(TIF)
After cross-linking, the FLAG-tagged p33 and FLAG-p92 were purified from solubilized membranous fraction of yeast extracts using a FLAG-affinity column. Top panel: Western blot analysis of the co-purified 6xHis-tagged Osh6p with anti-His antibody in the affinity-purified preparations. Middle panel: Western-blot analysis of the same samples as in the top panel, but using anti-FLAG antibody. Bottom panels: Western blot analysis of 6xHis-Osh6p with anti-His antibody in the total protein extract from yeast expressing the shown proteins. CB: Coomassie-stained SDS-PAGE of total protein extract. Each experiment was repeated two times.
(EPS)
The yeast cell replicating TBSV repRNA was imaged to detect the p33 replication protein via Alexa Flour 647 and actin filaments through staining with ATTO488-phalloidin. The bars represent 1 μm. The boxed area represents the 3D image to visualize the localization of actin and p33 replication protein in yeast. The image was prepared by a Nikon Super Resolution Microscope N-STORM and image processing was performed using NIS-element software.
(AVI)
Acknowledgments
We thank Dr. Judit Pogany for critical reading of the manuscript and Dr. Chingkai Chuang for very helpful suggestions. The authors thank the Nikon imaging team for their help with the super-resolution microscopy imaging. The COF1 mutant strains were kindly provided by Dr. David Drubin (UC Berkeley). Transgenic Nicotiana benthamiana plants expressing GFP-mTalin (mouse Talin) were a gift from Dr. Michael M. Goodin (University of Kentucky) and Nt-ADF2 clone was kindly provided by Dr. Alice Cheung (University of Massachusetts). Anti-p33 primary antibody was a gift from Dr. Herman B. Scholthof (Texas A&M University). The authors also thank Dr. Charles Boone (University of Toronto) for yeast ts mutants.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Foundation, MCB-1517751, awarded to PDN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. den Boon JA, Diaz A, Ahlquist P (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8: 77–85. 10.1016/j.chom.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A (2009) Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5: 318–328. 10.1016/j.chom.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 3. Nagy PD, Pogany J (2012) The dependence of viral RNA replication on co-opted host factors. Nature Reviews Microbiology 10: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu K, Nagy PD (2014) Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr Opin Virol 9C: 119–126. [DOI] [PubMed] [Google Scholar]
- 5. Belov GA, van Kuppeveld FJ (2012) (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol 2: 740–747. 10.1016/j.coviro.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romero-Brey I, Bartenschlager R (2014) Membranous replication factories induced by plus-strand RNA viruses. Viruses 6: 2826–2857. 10.3390/v6072826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K (2011) Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411: 374–382. 10.1016/j.virol.2010.12.061 [DOI] [PubMed] [Google Scholar]
- 8. Li Z, Nagy PD (2011) Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol 8: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagy PD, Pogany J (2010) Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv Virus Res 76: 123–177. 10.1016/S0065-3527(10)76004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hyodo K, Okuno T (2015) Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr Opin Virol 17: 11–18. 10.1016/j.coviro.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 11. Wang A (2015) Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol 53: 45–66. 10.1146/annurev-phyto-080614-120001 [DOI] [PubMed] [Google Scholar]
- 12. Nagy PD, Pogany J (2006) Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344: 211–220. [DOI] [PubMed] [Google Scholar]
- 13. White KA, Nagy PD (2004) Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol 78: 187–226. [DOI] [PubMed] [Google Scholar]
- 14. Panaviene Z, Panavas T, Serva S, Nagy PD (2004) Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol 78: 8254–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panavas T, Nagy PD (2003) Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314: 315–325. [DOI] [PubMed] [Google Scholar]
- 16. Nagy PD, Pogany J, Lin JY (2014) How yeast can be used as a genetic platform to explore virus-host interactions: from 'omics' to functional studies. Trends Microbiol 22: 309–316. 10.1016/j.tim.2014.02.003 [DOI] [PubMed] [Google Scholar]
- 17. Jonczyk M, Pathak KB, Sharma M, Nagy PD (2007) Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362: 320–330. [DOI] [PubMed] [Google Scholar]
- 18. Pogany J, White KA, Nagy PD (2005) Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol 79: 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panavas T, Hawkins CM, Panaviene Z, Nagy PD (2005) The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338: 81–95. [DOI] [PubMed] [Google Scholar]
- 20. Stork J, Kovalev N, Sasvari Z, Nagy PD (2011) RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409: 338–347. 10.1016/j.virol.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pogany J, Nagy PD (2008) Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol 82: 5967–5980. 10.1128/JVI.02737-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panaviene Z, Panavas T, Nagy PD (2005) Role of an internal and two 3'-terminal RNA elements in assembly of tombusvirus replicase. J Virol 79: 10608–10618. 16051853 [Google Scholar]
- 23. Pathak KB, Pogany J, Xu K, White KA, Nagy PD (2012) Defining the Roles of cis-Acting RNA Elements in Tombusvirus Replicase Assembly In Vitro. J Virol 86: 156–171. 10.1128/JVI.00404-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pogany J, Nagy PD (2015) Activation of Tomato Bushy Stunt Virus RNA-Dependent RNA Polymerase by Cellular Heat Shock Protein 70 Is Enhanced by Phospholipids In Vitro. J Virol 89: 5714–5723. 10.1128/JVI.03711-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagy PD (2008) Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol 46: 217–242. 10.1146/annurev.phyto.121407.093958 [DOI] [PubMed] [Google Scholar]
- 26. Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, et al. (2005) Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A 102: 10545–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panavas T, Serviene E, Brasher J, Nagy PD (2005) Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A 102: 7326–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD (2006) Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol 80: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD (2006) Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol 80: 7394–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mendu V, Chiu M, Barajas D, Li Z, Nagy PD (2010) Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 406: 342–351. 10.1016/j.virol.2010.07.022 [DOI] [PubMed] [Google Scholar]
- 31. Shah Nawaz-Ul-Rehman M, Martinez-Ochoa N, Pascal H, Sasvari Z, Herbst C, et al. (2012) Proteome-wide overexpression of host proteins for identification of factors affecting tombusvirus RNA replication: an inhibitory role of protein kinase C. J Virol 86: 9384–9395. 10.1128/JVI.00019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Pogany J, Panavas T, Xu K, Esposito AM, et al. (2009) Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385: 245–260. 10.1016/j.virol.2008.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serva S, Nagy PD (2006) Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80: 2162–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD (2008) Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol 82: 6911–6926. 10.1128/JVI.00702-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang RY, Nagy PD (2008) Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe 3: 178–187. 10.1016/j.chom.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 36. Sasvari Z, Izotova L, Kinzy TG, Nagy PD (2011) Synergistic Roles of Eukaryotic Translation Elongation Factors 1Bgamma and 1A in Stimulation of Tombusvirus Minus-Strand Synthesis. PLoS Pathog 7: e1002438 10.1371/journal.ppat.1002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang TS, Nagy PD (2011) Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant negative mutant of a host metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, in yeast and plants. J Virol 85: 9090–9102. 10.1128/JVI.00666-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, et al. (2010) Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog 6: e1001175 10.1371/journal.ppat.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barajas D, Jiang Y, Nagy PD (2009) A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog 5: e1000705 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang TS, Nagy PD (2011) Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant-negative mutant of a host metabolic enzyme, GAPDH, in yeast and plants. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang RY, Stork J, Nagy PD (2009) A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83: 3276–3287. 10.1128/JVI.02313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pogany J, Stork J, Li Z, Nagy PD (2008) In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A 105: 19956–19961. 10.1073/pnas.0810851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu K, Nagy PD (2015) RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc Natl Acad Sci U S A 112: E1782–E1791. 10.1073/pnas.1418971112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chuang C, Barajas D, Qin J, Nagy PD (2014) Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane. PLoS Pathog 10: e1003944 10.1371/journal.ppat.1003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barajas D, Xu K, de Castro Martin IF, Sasvari Z, Brandizzi F, et al. (2014) Co-opted Oxysterol-Binding ORP and VAP Proteins Channel Sterols to RNA Virus Replication Sites via Membrane Contact Sites. PLoS Pathog 10: e1004388 10.1371/journal.ppat.1004388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma M, Sasvari Z, Nagy PD (2011) Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology 415: 141–152. 10.1016/j.virol.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharma M, Sasvari Z, Nagy PD (2010) Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol 84: 2270–2281. 10.1128/JVI.02003-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah Nawaz-Ul-Rehman M, Reddisiva Prasanth K, Baker J, Nagy PD (2013) Yeast screens for host factors in positive-strand RNA virus replication based on a library of temperature-sensitive mutants. Methods 59: 207–216. 10.1016/j.ymeth.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 49. Lappalainen P, Drubin DG (1997) Cofilin promotes rapid actin filament turnover in vivo. Nature 388: 78–82. [DOI] [PubMed] [Google Scholar]
- 50. Okreglak V, Drubin DG (2007) Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol 178: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De La Cruz EM (2009) How cofilin severs an actin filament. Biophys Rev 1: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen Q, Pollard TD (2013) Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Curr Biol 23: 1154–1162. 10.1016/j.cub.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bamburg JR, Bernstein BW (2010) Roles of ADF/cofilin in actin polymerization and beyond. F1000 Biol Rep 2: 62 10.3410/B2-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bernstein BW, Bamburg JR (2010) ADF/cofilin: a functional node in cell biology. Trends Cell Biol 20: 187–195. 10.1016/j.tcb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moseley JB, Goode BL (2006) The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev 70: 605–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bamburg JR, Wiggan OP (2002) ADF/cofilin and actin dynamics in disease. Trends Cell Biol 12: 598–605. [DOI] [PubMed] [Google Scholar]
- 57. Bamburg JR, Bernstein BW (2008) ADF/cofilin. Curr Biol 18: R273–275. 10.1016/j.cub.2008.02.002 [DOI] [PubMed] [Google Scholar]
- 58. Mishra M, Huang J, Balasubramanian MK (2014) The yeast actin cytoskeleton. FEMS Microbiol Rev 38: 213–227. 10.1111/1574-6976.12064 [DOI] [PubMed] [Google Scholar]
- 59. Taylor MP, Koyuncu OO, Enquist LW (2011) Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol 9: 427–439. 10.1038/nrmicro2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44. [DOI] [PubMed] [Google Scholar]
- 61. Kovalev N, Pogany J, Nagy PD (2014) Template role of double-stranded RNA in tombusvirus replication. J Virol 88: 5638–5651. 10.1128/JVI.03842-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bowman GD, Nodelman IM, Hong Y, Chua NH, Lindberg U, et al. (2000) A comparative structural analysis of the ADF/cofilin family. Proteins 41: 374–384. [DOI] [PubMed] [Google Scholar]
- 63. McCurdy DW, Kovar DR, Staiger CJ (2001) Actin and actin-binding proteins in higher plants. Protoplasma 215: 89–104. [DOI] [PubMed] [Google Scholar]
- 64. Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, et al. (2002) The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barajas D, Xu K, Sharma M, Wu CY, Nagy PD (2014) Tombusviruses upregulate phospholipid biosynthesis via interaction between p33 replication protein and yeast lipid sensor proteins during virus replication in yeast. Virology 471-473C: 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG (1997) Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J 16: 5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, et al. (2011) Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29: 361–367. 10.1038/nbt.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vidali L, McKenna ST, Hepler PK (2001) Actin polymerization is essential for pollen tube growth. Mol Biol Cell 12: 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kwon KW, Park H, Doh J (2013) Migration of T cells on surfaces containing complex nanotopography. PLoS One 8: e73960 10.1371/journal.pone.0073960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401. [DOI] [PubMed] [Google Scholar]
- 71. Barajas D, Martin IF, Pogany J, Risco C, Nagy PD (2014) Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Tomato Bushy Stunt Virus Replicase. PLoS Pathog 10: e1004087 10.1371/journal.ppat.1004087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peremyslov VV, Cole RA, Fowler JE, Dolja VV (2015) Myosin-Powered Membrane Compartment Drives Cytoplasmic Streaming, Cell Expansion and Plant Development. PLoS One 10: e0139331 10.1371/journal.pone.0139331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Avisar D, Prokhnevsky AI, Dolja VV (2008) Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82: 2836–2843. 10.1128/JVI.02246-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. de Castro IF, Volonte L, Risco C (2013) Virus factories: biogenesis and structural design. Cell Microbiol 15: 24–34. 10.1111/cmi.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Novick P, Botstein D (1985) Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell 40: 405–416. [DOI] [PubMed] [Google Scholar]
- 76. Henty-Ridilla JL, Shimono M, Li J, Chang JH, Day B, et al. (2013) The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog 9: e1003290 10.1371/journal.ppat.1003290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sasvari Z, Nagy PD (2010) Making of Viral Replication Organelles by Remodeling Interior Membranes. Viruses-Basel 2: 2436–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pathak KB, Sasvari Z, Nagy PD (2008) The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379: 294–305. 10.1016/j.virol.2008.06.044 [DOI] [PubMed] [Google Scholar]
- 79. Sasvari Z, Alatriste Gonzalez P, Nagy PD (2014) Tombusvirus-yeast interactions identify conserved cell-intrinsic viral restriction factors. Front Plant Sci 5: 383 10.3389/fpls.2014.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sattlegger E, Chernova TA, Gogoi NM, Pillai IV, Chernoff YO, et al. (2014) Yeast studies reveal moonlighting functions of the ancient actin cytoskeleton. IUBMB Life 66: 538–545. 10.1002/iub.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Z, Gonzalez PA, Sasvari Z, Kinzy TG, Nagy PD (2014) Methylation of translation elongation factor 1A by the METTL10-like See1 methyltransferase facilitates tombusvirus replication in yeast and plants. Virology 448C: 43–54. [DOI] [PubMed] [Google Scholar]
- 82. Stolp B, Reichman-Fried M, Abraham L, Pan X, Giese SI, et al. (2009) HIV-1 Nef interferes with host cell motility by deregulation of Cofilin. Cell Host Microbe 6: 174–186. 10.1016/j.chom.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 83. Yoder A, Yu D, Dong L, Iyer SR, Xu X, et al. (2008) HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134: 782–792. 10.1016/j.cell.2008.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Le Clainche C, Drubin DG (2004) Actin lessons from pathogens. Mol Cell 13: 453–454. [DOI] [PubMed] [Google Scholar]
- 85. Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, et al. (2012) The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol 158: 1359–1370. 10.1104/pp.111.189605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Harries PA, Park JW, Sasaki N, Ballard KD, Maule AJ, et al. (2009) Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci U S A 106: 17594–17599. 10.1073/pnas.0909239106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Panaviene Z, Nagy PD (2003) Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology 317: 359–372. [DOI] [PubMed] [Google Scholar]
- 88. Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, et al. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162. 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
- 89. Xu K, Huang TS, Nagy PD (2012) Authentic in vitro replication of two tombusviruses in isolated mitochondrial and endoplasmic reticulum membranes. J Virol 86: 12779–12794. 10.1128/JVI.00973-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(A) Accumulation of TBSV repRNA in cof1-5 ts or wt yeast at the semi-permissive (32°C) temperature. To launch TBSV repRNA replication, we expressed His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter in the parental (BY4741) and cof1-5 ts yeast strains. Northern blot analysis was used to detect DI-72(+) repRNA accumulation, which was normalized based on 5S rRNA. Each experiment was repeated three times. (B) Testing Cof1p mutants defective in Act1p binding for their ability to inhibit TBSV repRNA accumulation in yeast. (C) Testing Cof1p mutants that bind Act1p for their ability to inhibit TBSV repRNA accumulation in yeast. See further details in panel A. (D) Cof1p mutant proteins interact with the TBSV p33 replication protein in yeast. The split ubiquitin assay was used to test binding between p33 and Cof1p in yeast. The bait p33 was co-expressed with the shown prey proteins. SSA1 and the empty prey vector (NubG-X) were used as positive and negative controls, respectively. The experiment with wt Cof1p is encircled.
(EPS)
(A-B) The wt or cof1-8 ts yeast co-expressed the His6-p33 and His6-p92 from the copper-inducible CUP1 promoter and TBSV DI-72(+) repRNA from the constitutive ADH1 promoter. Over-expression of the untagged native Cof1p was done from the GAL1 promoter. Top images: Northern blot analysis of TBSV repRNA in yeast samples over-expressing Cof1p or without over-expression is shown. repRNA replication took place for 24 hours at 23°C or 32°C in wt or cof1-8 ts yeast before RNA analysis. The accumulation level of DI-72(+) repRNA (shown in percentage) was normalized based on 18S rRNA.
(EPS)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing p33-RFP and infected with TBSV using sap inoculation. Note the localization of large p33-RFP containing areas (i.e., replication organelles) frequently at the intersection of actin cables. The bars represent 20 μm.
(TIF)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with a plasmid expressing p36-RFP and infected with CIRV using sap inoculation. Note the localization of large p36-RFP containing areas frequently at the intersection of actin cables. The bars represent 20 μm.
(TIF)
The GFP-mTalin transgenic N. benthamiana plants were agro-infiltrated with an empty pGD plasmid. The bars represent 20 μm.
(TIF)
After cross-linking, the FLAG-tagged p33 and FLAG-p92 were purified from solubilized membranous fraction of yeast extracts using a FLAG-affinity column. Top panel: Western blot analysis of the co-purified 6xHis-tagged Osh6p with anti-His antibody in the affinity-purified preparations. Middle panel: Western-blot analysis of the same samples as in the top panel, but using anti-FLAG antibody. Bottom panels: Western blot analysis of 6xHis-Osh6p with anti-His antibody in the total protein extract from yeast expressing the shown proteins. CB: Coomassie-stained SDS-PAGE of total protein extract. Each experiment was repeated two times.
(EPS)
The yeast cell replicating TBSV repRNA was imaged to detect the p33 replication protein via Alexa Flour 647 and actin filaments through staining with ATTO488-phalloidin. The bars represent 1 μm. The boxed area represents the 3D image to visualize the localization of actin and p33 replication protein in yeast. The image was prepared by a Nikon Super Resolution Microscope N-STORM and image processing was performed using NIS-element software.
(AVI)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.