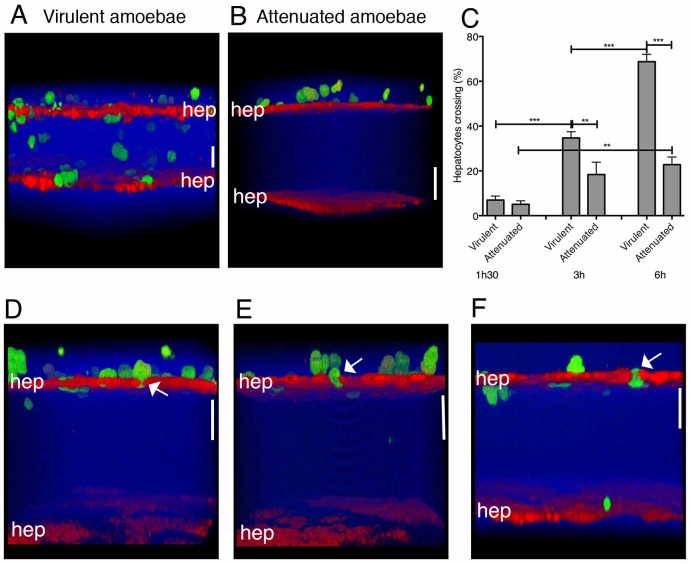
Fig 4. 3D liver model with the double hepatocyte layer: visualization and quantification of hepatocyte crossing by E. histolytica.
For two-photon microscopy acquisitions, hepatocytes were labelled with red and amoebae with green cell tracker. Col-I fibres were visualized by SHG signal (in blue). (A-B) 3D transversal reconstruction (ICY software) of 3D liver model setup 2 after 3h of interaction with virulent (A) or virulence-attenuated (B) trophozoites. (C) Quantification of the hepatocyte crossing by E. histolytica after 1.5h, 3h and 6h of interaction with the model. Obtained from 3 independent experiments. Standard deviation and statistical significance indicated, with ** = p < 0.01 and *** = p < 0.001. D-F) 3D transversal reconstruction (ICY software) enabling visualization of amoebae squeezing in between hepatocytes during hepatic crossing at 1.5h, 3h and 6h of interaction with the model (arrows). Scale bar = 50μm.