Figure 6.
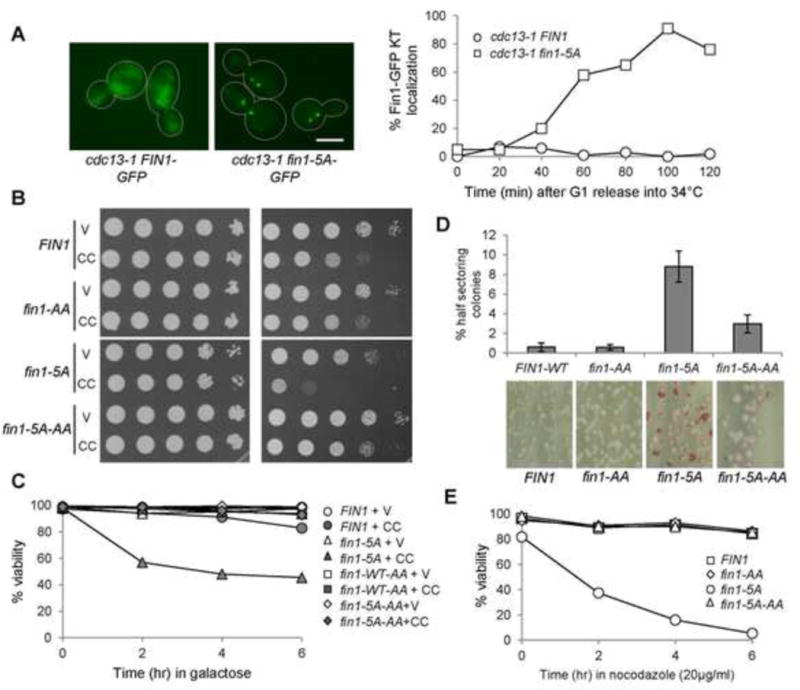
Nonphosphorylatable Fin1-5A show premature kinetochore localization and fin1-5A mutants are sensitivity to CIK1-CC overexpression. (A) Fin1-5A-GFP prematurely localizes at the kinetochore in cdc13-1 arrested cells. cdc13-1 fin1Δ cells with Fin1-GFP or Fin1-5A-GFP were synchronized in G1 at 25°C and then released into 34° YPD medium. Cells were collected over time and fixed. The percentage of cells with two clear GFP dots was counted (n >100). The localization of Fin1-GFP and Fin1-5A-GFP in some cdc13-1-arrested cells is shown in the left. Scale bar, 5μm. (B) fin1-5A mutants show PP1-dependent sensitivity to CIK1-CC overexpression. Saturated cells with indicated genotypes were 10-fold serial diluted, spotted onto glucose and galactose plates, and then incubated at 30°C for 2 days before scanning. fin1-5A: 5 CDK phosphorylation sites are mutated. fin1-5A-AA: both CDK phosphorylation sites and PP1 binding motif are mutated. V (vector), CC (PGALCIK1-CC). (C) fin1-5A cells overexpressing CIK1-CC show viability loss, which depends on Fin1’s PP1 binding. Log-phase cells in raffinose were released into 2% galactose medium. Samples were taken every 2 hr and spread onto YPD plates to determine plating efficiency (n > 300). (D) fin1-5A mutant cells show elevated chromosome loss, which depends on its PP1 binding. Cells harboring chromosome fragment III (CFIII) with indicated genotypes were incubated in URA dropout medium and then spread onto URA+ plates lacking adenine. The percentage of half-red colonies was counted after incubation for 3 days at 25°C. Here shows the average of three independent experiments (n > 400). (E) fin1-5A mutant cells show PP1-binding dependent sensitivity to nocodazole exposure. Cells with the indicated genotypes were released into YPD containing 20μg/ml of nocodazole. Cells were collected every 2 hr and spread onto YPD plates to determine the viability.
