Abstract
Adenosine triphosphate (ATP) acts on P2X receptors to initiate signal transmission. P2X7 receptors play a role in the pathophysiological process of myocardial ischemic injury. Long noncoding RNAs (lncRNAs) participate in numerous biological functions independent of protein translation. LncRNAs are implicated in nervous system diseases. This study investigated the effects of NONRATT021972 small interference RNA (siRNA) on the pathophysiologic processes mediated by P2X7 receptors in stellate ganglia (SG) after myocardial ischemic injury. Our results demonstrated that the expression of NONRATT021972 in SG was significantly higher in the myocardial ischemic (MI) group than in the control group. Treatment of MI rats with NONRATT021972 siRNA, the P2X7 antagonist brilliant blue G (BBG), or P2X7 siRNA improved the histology of injured ischemic cardiac tissues and decreased the elevated concentrations of serum myocardial enzymes, creatine kinase (CK), CK isoform MB (CK-MB), lactate dehydrogenase (LDH) and aspartate aminotransferase (AST) compared to the MI rats. NONRATT021972 siRNA, BBG, or P2X7 siRNA treatment in MI rats decreased the expression levels of P2X7 immunoreactivity, P2X7 messenger RNA (mRNA), and P2X7 protein, interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), and phosphorylated p38 mitogen-activated protein kinase (p38 MAPK) in the SG compared to MI rats. NONRATT021972 siRNA treatment prevented the pathophysiologic processes mediated by P2X7 receptors in the SG after myocardial ischemic injury.
Keywords: Stellate ganglia, Adenosine triphosphate, P2X7 receptor, Myocardial ischemia, Long noncoding RNA
Introduction
Long noncoding RNAs (lncRNAs) are transcripts greater than 200 nucleotides in length [1, 2]. LncRNAs were initially considered to be byproducts of RNA polymerase II transcription without a biological function. However, the role of increasing numbers of lncRNAs in a variety of biological functions independent of protein translation was demonstrated [3, 4]. LncRNAs participate in posttranscriptional gene regulation via the control of protein synthesis, RNA maturation and transport, and transcriptional gene silencing via regulation of chromatin structure [3–6]. LncRNAs, unlike proteins, act in cis at the site of transcription (act locally to regulate the expression of nearby genes) or trans (distally to regulate gene expression across multiple chromosomes) [7]. LncRNAs allow cells to address regional-, locus-, or allele-specific transcriptions [8]. LncRNAs impact the formation of nuclear domains at the regional level and the transcriptional status of an entire chromosome. LncRNAs also participate in the interaction of two different chromosomal regions. LncRNAs regulate the chromatin state and the activity of a chromosomal locus or specific gene at a locus-specific level [7]. LncRNAs are implicated in the processes of many pathological conditions [9].
The native agonist extracellular adenosine 5′-triphosphate (ATP) acts at P2X receptors on the plasma membrane. P2X ionotropic ligand-gated ion channel receptors are classified into P2X1–P2X7 [10–13]. P2X receptors participate in the regulation of cardiovascular function and disease [14, 15]. P2X7 receptors play important roles in the transmission of inflammatory and nociceptive signals [11, 12, 16–18]. Cervical sympathetic ganglia are involved in ischemic heart diseases [19–29]. The ATP that is released from injured cells following myocardial ischemic injury activates P2X receptors in sympathetic afferent and efferent nerves [11, 19–21, 23, 26–31]. Our previous studies implicated P2X7 receptors in cervical sympathetic ganglia in myocardial ischemia [19, 23, 27]. LncRNAs are involved in normal development and disease [32]. However, the roles of lncRNAs in sympathetic ganglia are poorly characterized. NONRATT021972 is an lncRNA [http://www.noncode.org; 33]. This research investigated the effects of lncRNA NONRATT021972 small interference RNA (siRNA) on the pathophysiologic processes mediated by P2X7 receptors in the stellate ganglia (SG) after myocardial ischemic injury.
Materials and methods
Sprague-Dawley (SD) rats weighing 180–220 g were used in all experiments. The Animal Care and Use Committee of Medical College of Nanchang University reviewed and approved the use of animals. Experiments were performed in accordance with the guidelines of the US National Institutes of Health for the care and use of animals for experimental procedures. Rats were randomly divided into seven groups (eight rats per group): control group (Con group), sham group, myocardial ischemic group (MI group), myocardial ischemic rats treated with the P2X7 receptor antagonist brilliant blue G (BBG) group (MI + BBG group), myocardial ischemic group rats treated with NONRATT021972 siRNA group (MI + NONRATT021972 si), myocardial ischemic rats treated with P2X7 siRNA group (MI + P2X7 si), and myocardial ischemic rats transfected with scrambled siRAN group (MI + SC si group). The myocardial ischemic model was established via occlusion of the left coronary artery (LCA) [18]. Briefly, rats were anesthetized with 10 % chloral hydrate (0.3 mL/100 g body weight) via intraperitoneal injection. Rats were mechanically ventilated, and a thoracotomy was performed. A 5-0 suture on a small, curved needle was passed through the myocardium beneath the LCA. The suture was pulled tightly against a vinyl tube to ligate the blood vessel. The LCA was not ligated in the sham surgery, but the cardiac pericardium was damaged.
Electrocardiogram (ECG) changes were continuously monitored post-surgery. The relative voltage of the negative peak of the S-wave against the QQ line was defined as the ST segment. Myocardial ischemia was confirmed as an observed abnormal Q wave and ST-segment displacement in a lead II ECG. Rats in the MI + BBG group received intraperitoneal BBG (30 mg/kg, dissolved in saline at a final concentration of 0.2 mg/mL) daily for 30 days beginning 24 h after surgery [34]. Rats in the MI + NONRATT021972 si, MI + P2X7 si, and MI + SC si groups received NONRATT021972 si, P2X7 si, and scrambled siRAN, respectively, via sublingual injections.
The following small interfering RNA target sequences were used:
- P2X7 siRNA
GTGCAGTGAATGAGTACTA.
- NONRATT021972
TGTGAATCATGGAAATATC.
Reverse transcription-polymerase chain reaction (RT-PCR)
Serum RNA from patients with coronary heart disease or control healthy subjects was tested using RT-PCR. Total RNA was isolated from sera using the RNApure Circulating Reagent (CWBIO, Beijing, cat.no.CW2281), with some modifications. Briefly, 300 μL of serum was added to three volumes of RNApure Circulating Reagent, mixed thoroughly by vortexing and left at room temperature for 5 min. A 1/5 volume of chloroform was added, shaken vigorously for 30 s, incubated for 5 min at room temperature, and centrifuged for 20 min at 12,000×g at 4 °C. The upper aqueous phase was transferred to a new collection tube, and 1 volume of isopropanol was added, mixed thoroughly for 30 min at room temperature and centrifuged for 20 min at 12,000×g at 4 °C. The precipitate was washed twice with 1 mL of 75 % ethanol which is diluted with diethyl pyrocarbonate (DEPC) water. RNA was eluted by 20 μL of RNase-free water. RNA quality was profiled using a NanoDrop 2000 (Thermo scientific, USA).
Total RNA (1000 ng) was used as a template for reverse transcription using the Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA). PCR amplification of NONRATT021972 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, control) was performed according to our previous method using oligonucleotides as described previously [35]. The following primers were used: NONRATT021972, sense 5′-CGGGATCCAGGCCTGCTGAAAATGACTGAGTATAAAC-3′ and antisense 5′-CCGCTCGAGTTCATAGTCACCATAACTATTTTTATTACATTAC-3′; and GAPDH sense 5′-CAGGGCTGCTTTTAACTCTGGT-3′ and antisense 5′-GATTTTGGAGGGATCTCGCT-3′. The length of the PCR product was 250 bp for NONRATT021972 and 199 bp for GAPDH. PCR conditions of NONRATT021972 and GAPDH included a hotstart at 94 °C for 3 min, 45-s denaturation (94 °C), 45-s annealing (57 °C), 45-s extension (72 °C) for 36 or 32 amplification cycles, and a 5-min final extension at 72 °C. PCR products were separated on 1.5 % agarose gels and stained with ethidium bromide (EB) using standard protocols. These results are expressed as the ratio of the NONRATT021972 band intensity/GAPDH band intensity. Data were analyzed using Image-Pro Plus software.
Quantitative real-time PCR
Total RNA was isolated from SG using the TRIzol Total RNA Reagent (Beijing Tiangen Biotech Co.). The reverse transcription reaction was completed using a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Glen Bernie, USA) following the manufacturer’s instructions. The primers were designed using Primer Express 3.0 software (Applied Biosystems), and the following sequences were used: P2X7, sense 5′-CTTCGGCGTGCGTTTTG-3′ and antisense 5′-AGGACAGGGTGGATCCAATG-3′; NONRATT021972, sense 5′-TAGGATGAGTACCAGTCAGGT-3′ and antisense 5′-TTTTTGGTTTTTTGACAGGG-3′; and β-actin, sense 5′-GCTCTTTTCCAGCCTTCCTT-3′ and antisense 5′-CTTCTGCATCCTGTCAGCAA-3′. Quantitative PCR was performed using the SYBR® Green MasterMix in an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems Inc., Foster City, CA). The thermal cycling parameters were 95 °C for 30 s, followed by 40 cycles of amplifications at 95 °C for 5 s, 60 °C for 30 s [36]. Amplification specificity was determined using a melting curve, and the results were processed using software within the ABI7500 PCR instrument.
ISH
Messenger RNA (mRNA) expression was assessed using in situ hybridization (ISH) [20]. Rats were anesthetized, and the SG were dissected immediately. SG were fixed in 4 % paraformaldehyde (PFA) for 2 h at room temperature and transferred to 15 % sucrose in 4 % PFA overnight. Tissues were sectioned at 15 μm. Samples were stored in 4 % PFA in a cryostat at 4 °C. DEPC water was used for all solutions and appliances of ISH. An ISH kit for NONRATT021972 was used. Sections were treated with 0.05 % H2O2, followed by digestion with pepsin at 37 °C for 1–2 min. The reaction was terminated with 0.5 mol phosphate-buffered saline (PBS), and sections were washed with PBS for 15 min. Sections were incubated in a prehybridization buffer for 2 h at 37 °C and hybridization buffer overnight at 37 °C. Sections were washed with a gradient of SSC (2× SSC 17.6 g sodium chloride, 8.8 g sodium citrate in 1000 mL distilled water) thoroughly, 2× SSC for 10 min, 0.5× SSC for 15 min, and 0.2× SSC for 15 min to remove background signals. Sections were treated with a biotinylated digoxin antibody at 37 °C for 2 h. Sections were thoroughly washed with PBS and incubated with strept avidin-biotin complex for 30 min and biotinylated peroxidase for 30 min at 37 °C. The color reaction was developed using a DAB substrate, and sections were dehydrated and mounted using neutral gum.
HE staining of myocardial tissue
Myocardial tissues were washed in PBS, fixed with 4 % PFA for 24 h and dehydrated in 30 % sucrose overnight at 4 °C. Myocardial tissues were cut into 14-μm-thick sections using a cryostat. The sections were stained with hematoxylin for 20 s, washed with tap water for 10 min, and dehydrated in 70, 80, and 90 % ethanol for 2 min each. The sections were stained with eosin for 15 s and dehydrated in 95, 95, 100, and 100 % ethanol for 2 min each.
Immunohistochemistry
The rats were killed 30 days after surgery. The heart and SG tissues were dissected immediately and washed in PBS. Tissue samples were fixed in 4 % PFA for 24 h, and the ganglia were dehydrated in 30 % sucrose overnight at 4 °C. The ganglia were cut into 12-μm-thick sections on a cryostat. Sections were washed with PBS three times and incubated in 3 % H2O2 for 7 min to block endogenous peroxidase activity. The sections were washed in PBS three times and incubated in 5 % bovine serum albumin (BSA) for 2 h at 37 °C. Sections were rinsed three times in PBS and incubated with a rabbit anti-P2X7 antibody (1:100; Alomone Labs, Jerusalem, Israel), rabbit anti-tumor necrosis factor-α (TNF-α) antibody (1:100 dilution, Millipore International Inc., USA), and rabbit anti-interleukin-6 (IL-6) antibody (1:100 dilution, Abcam International Inc., USA), diluted in PBS overnight at 4 °C. Sections were rinsed three times in PBS and incubated with a biotinylated goat anti-rabbit secondary antibody (Beijing Zhongshan Biotech, Co.) for 1 h at 37 °C. The preparations were washed in PBS and incubated with streptavidin-horseradish peroxidase (Beijing Zhongshan Biotech, Co.) for 30 min at room temperature. Staining was developed using diaminobenzidine chromogen for 2 min, and the slides were washed with distilled water and cover-slipped. The sections were mounted and examined under a microscope (Olympus TH4-200, Japan). Changes in integrated optical density (IOD) for ganglionic neurons were analyzed using Image Pro-Plus software.
ELISA measurement of serum cardiac enzymes and cytokine levels
Blood samples were collected via orbit vein 30 days after surgery. Samples were maintained at room temperature for 30 min and centrifuged at 3000 rpm for 10 min. Sera were collected and stored at −20 °C. Levels of cardiac enzymes in sera, including CK CK-MB, LDH, and AST, were measured using an automatic electrochemiluminescence immunoassay analyzer (Roche COBAS E601, USA) [37]. Serum cytokine levels, including TNF-αand IL-6, were quantified using an enzyme-linked immunosorbent assay (ELISA) kit [20] following the manufacturer’s protocol (Senxiong Co., Shanghai, China). The reactions were read using an ELISA reader (Rayto, RT-6000, USA) at 450 nm.
Western blotting
Protein expression levels were determined using Western blotting [20, 37, 38]. Rats were killed 30 days after surgery. Ganglia tissues were dissected and flushed with ice-cold PBS. Isolated tissues were homogenized via mechanical disruption in lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.1 % dodecyl sodium sulfate, 1 % Nonidet P-40, 0.5 % sodium deoxycholate, 100 μg/mL phenylmethylsulfonyl fluoride, and 1 μg/mL aprotinin). The homogenate was incubated on ice for 40 min and centrifuged at 12,000 rpm for 10 min. The supernatant was collected, and protein concentration was quantified using the Lowry method. Samples were diluted with loading buffer (100 mM Tris-Cl, 200 mM dithiothreitol, 4 % sodium deodecylsulfate, 0.2 % bromophenol blue, and 20 % glycerol) and heated at 95 °C for 5 min. Equal amounts of total protein (20 μg) were separated using SDS-polyacrylamide gel electrophoresis in a Bio-Rad electrophoresis and blotting system. Separated proteins were transferred to PVDF membranes using the same system. The membranes were blocked with 5 % nonfat dry milk in a solution of 25 mM Tris-buffered saline (pH 7.2) plus 0.1 % Tween 20 (TBST) for 2 h at room temperature and incubated with a rabbit anti-P2X7 antibody (1:1000 dilutions, Alomone Labs, Jerusalem, Israel), mouse monoclonal anti-β-actin antibody (1:800 dilution, Advanced Immunochemicals, Long Beach, CA), rabbit anti-p-P38 antibody (1:1000 dilutions), and rabbit anti-P38 antibody (1:1000), at 4 °C overnight. Membranes were washed in TBST and incubated with HRP-conjugated IgG (1:5000 dilutions, Beijing Zhongshan Biotechnology Co.) for 1 h at room temperature. Membranes were washed in TBST, and protein bands were developed using an enhanced chemiluminescence (ECL) kit (Shanghai Pufei Biotechnology Co.) according to the manufacturer’s protocol. Chemiluminescent signals were collected on autoradiography films. Band intensity was quantified using Image Pro-Plus software. The relative band intensity of target proteins was normalized against the intensity of respective β-actin bands as internal controls.
Statistical analysis
Data were analyzed using SPSS 11.5 software. Numerical values are reported as the means ± SE. Statistical significance was determined using one-way analysis of variance (ANOVA) followed by Fisher’s post hoc test for multiple comparisons. A p < 0.05 was considered statistically significant.
Results
Changes in NONRATT021972 expression in SG
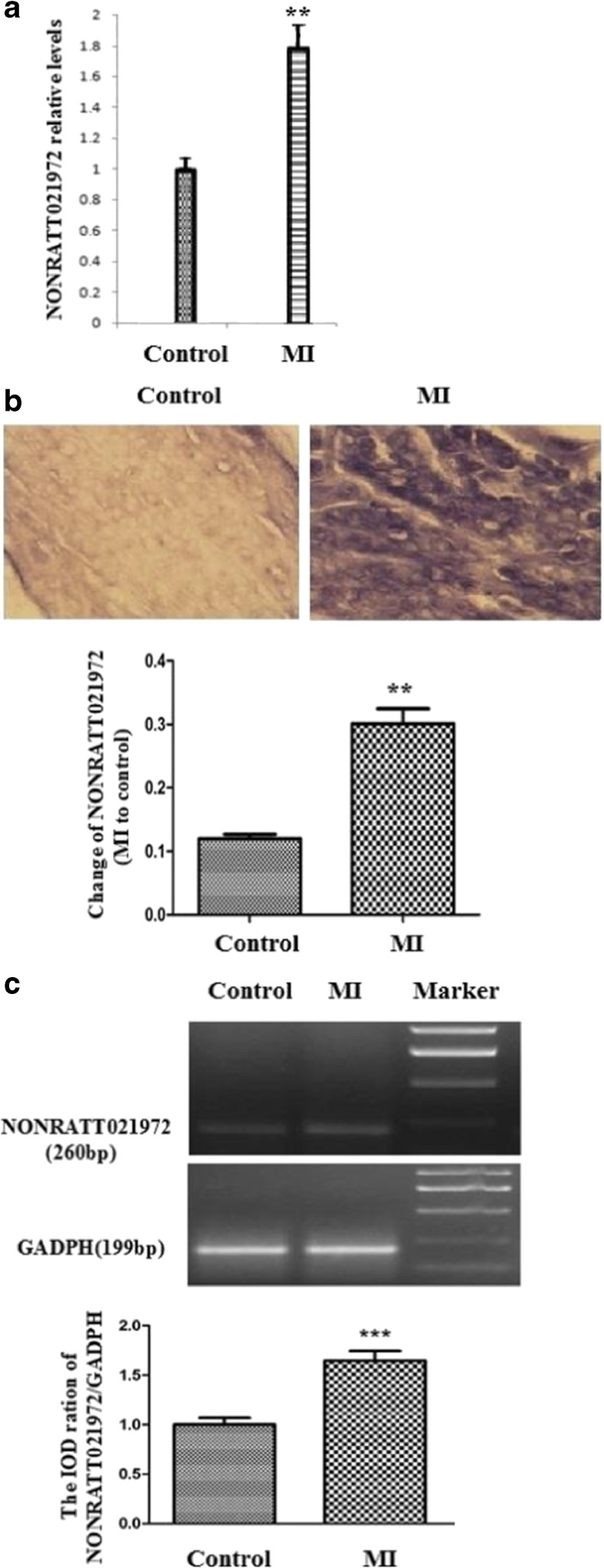
NONRATT021972 expression in SG was examined using real-time PCR, and NONRATT021972 expression was significantly higher in the MI group than the control group (p < 0.01) (Fig. 1a). ISH demonstrated that NONRATT021972 expression in SG was significantly higher in the MI group than the control group (p < 0.01) (Fig. 1b). Serum NONRATT021972 concentrations were higher in patients with coronary heart disease compared to control healthy subjects (p < 0.05, n = 8 for each group, Fig. 1c).
Fig. 1.
Expression levels of NONRATT021972 in SG. a The histogram of relative mRNA levels. The expression levels of NONRATT021972 in SG were significantly higher in the MI group than the Con group (p < 0.01). , n = 3. **p < 0.01 compared to the Con group. b The expression of NONRATT021972 in SG is significantly higher in the MI group than the control group assessed using ISH (p < 0.01). c Serum NONRATT021972 concentrations in patients with coronary heart disease were higher compared to control healthy subjects as assessed using RT-PCR (p < 0.05, n = 8 for each group)
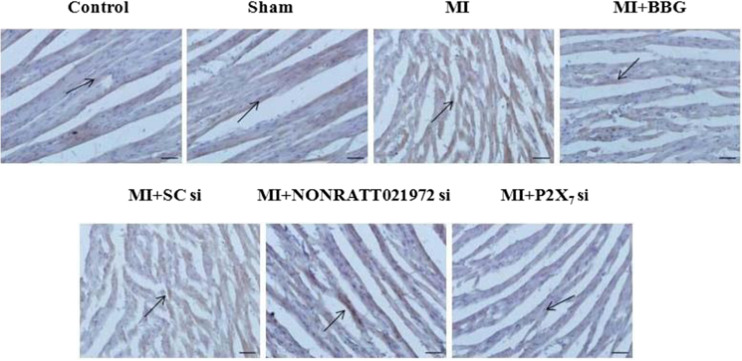
The effects of NONRATT021972 siRNA on myocardial structure and serum myocardial enzymes
Hematoxylin-eosin (HE) staining measured structural changes in myocardial tissue. The HE stain of cardiac tissue in MI rats revealed solidified necrosis of myocardial fibers, nuclei fragmentation and disappearance, an irregular coarse granular cytoplasm, interstitial edema, and a small amount of neutrophil infiltration. Myocardial fibers were pink in the Con and Sham groups, and the cytoplasm and nucleus were clear. The cell membrane exhibited good integrity, and the cellular arrangement was orderly with moderate intercellular spacing. Solidified necrosis of myocardial fibers, nuclei fragmentation, and disappearance in MI rats treated with BBG, NONRATT021972si, or P2X7 si were ameliorated in comparison with those in MI rats. The injury changes in cardiac tissues in the MI + BBG, MI+ NONRATT021972si, and MI + P2X7 si groups improved (Fig. 2).
Fig. 2.
Changes in HE staining in cardiac tissues of each group of rats. HE stains of cardiac tissue in myocardial ischemic rats show solidified necrosis of myocardial fibers, nuclei fragmentation and disappearance, an irregular coarse granular cytoplasm, interstitial edema, and a small amount of neutrophil infiltration. In the Con group and Sham group, the myocardial fibers were pink, and the cytoplasm and nucleus were clear. The cell membrane exhibited integrity, and cellular arrangement was regular with moderate intercellular space. Solidified necrosis of myocardial fibers, nuclei fragmentation, and disappearance in MI rats treated with BBG, NONRATT021972si, or P2X7 si were ameliorated in comparison with those in MI rats. The injury changes in cardiac tissues in the MI + BBG, MI+ NONRATT021972si, and MI + P2X7 si groups improved. Scale bar at 50 μm
Changes in myocardial enzymes were measured using an automatic electrochemiluminescence immunoassay analyzer. Serum concentrations of CK-MB, CK, LDH, and AST were significantly higher in the MI group than the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (n = 8) (p < 0.01, F (CK-MB)(5,42) = 3.76, F (CK)(5,42) = 10.23, F (LDH)(5,42) = 7.14, F (AST)(5,42) = 5.34). There were no differences between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (CK-MB)(4,35) = 1.03, F (CK)(4,35) = 1.46, F (LDH)(4,35) = 1.57, F (AST)(4,35) = 2.04). No differences were observed between the same MI and MI + SC si groups (p > 0.05) (Table 1).
Table 1.
The concentration of CK-MB, CK, LDH, and AST in the blood serum of the each group rats
| Groups | CK-MB (U/L) | CK (U/L) | LDH (U/L) | AST (U/L) |
|---|---|---|---|---|
| Con | 41.61 ± 3.82 | 327.15 ± 30.24 | 290.27 ± 21.14 | 78.78 ± 8.34 |
| Sham | 29.8 ± 2.53 | 259.57 ± 25.36 | 311.43 ± 27.50 | 84.15 ± 8.20 |
| MI | 97.50 ± 8.45** | 1852.44 ± 180.50** | 689.16 ± 68.34** | 197.25 ± 14.67** |
| MI + BBG | 43.13 ± 6.58 | 261.51 ± 28.33 | 321.41 ± 36.62 | 54.88 ± 5.94 |
| MI + SCsi | 85.45 ± 6.35** | 2103.47 ± 213.48** | 723.89 ± 72.58** | 223.63 ± 21.59** |
| MI + NONRATT021972si | 32.53 ± 2.76 | 294.18 ± 23.66 | 307.46 ± 30.84 | 57.22 ± 53.75 |
| MI + P2X7 si | 35.11 ± 4.21 | 347.43 ± 35.89 | 340.24 ± 35.54 | 74.19 ± 71.01 |
The serum concentration of CK-MB, CK, LDH, and AST in MI group was significantly higher than those in Con group, Sham group, MI + BBG group, MI+ NONRATT021972 si group, or MI + P2X7 si group (p < 0.01). There were no differences among Con, Sham, MI + BBG, MI+ NONRATT021972 si, and MI + P2X7 si groups (p > 0.05). It was the same between MI group and MI + SC si group (p > 0.05). , n = 8
**p < 0.01 compared with Con group
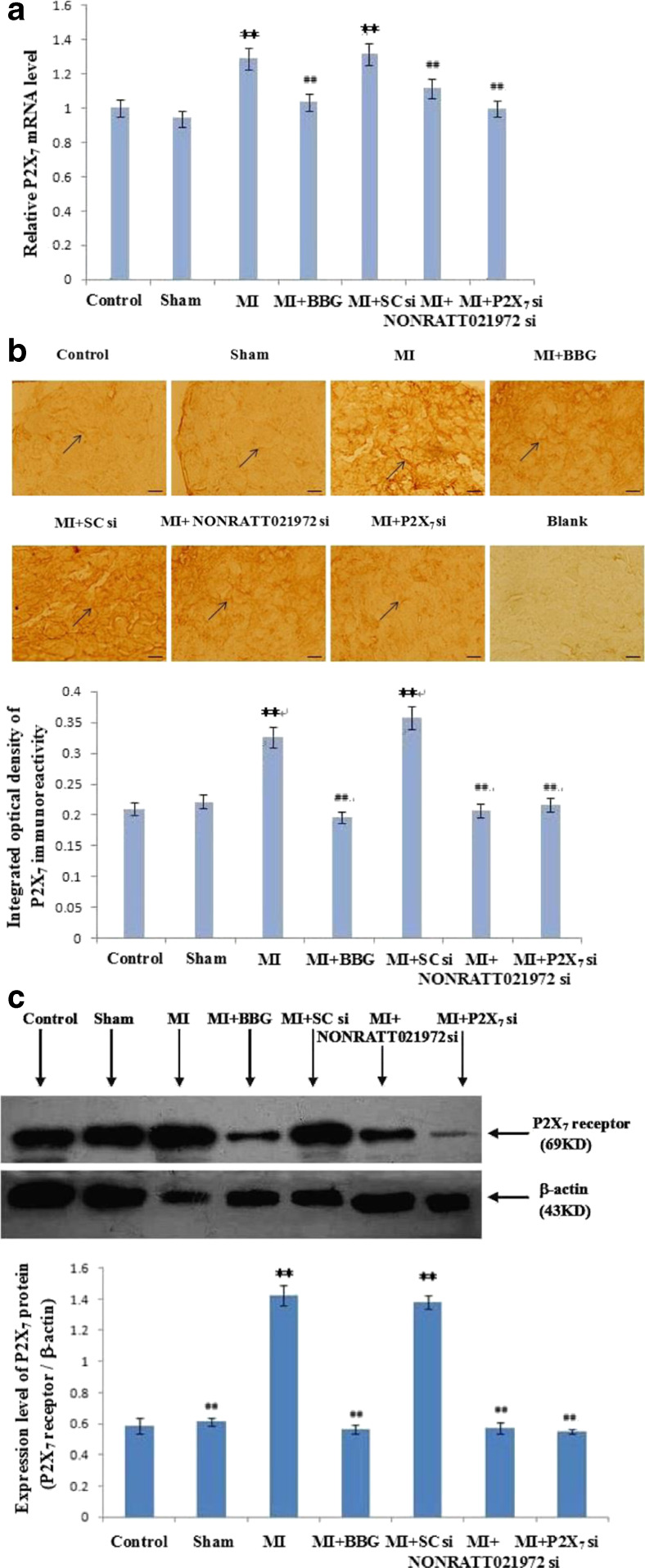
The effects of NONRATT021972 siRNA on the expression of P2X7 mRNA, immunoreactivity, and protein in SG
The expression level of P2X7 mRNA in SG was measured using quantitative real-time PCR. P2X7 mRNA expression in the MI and MI + SC si groups was significantly higher than the Con and Sham groups. P2X7 mRNA expression in the MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups decreased significantly compared to the MI group (p < 0.01, F (5,12) = 11.23). No difference was found between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F(4,10) = 2.19). No difference was found between the MI and MI + SC si groups (p > 0.05) (Fig. 3a).
Fig. 3.
The expression levels of P2X7 receptor mRNA, immunohistochemistry, and protein in SG of each group. a The expression levels of P2X7 receptor mRNA in SG were measured using quantitative real-time PCR. Histogram shows the relative mRNA levels. , n = 3. **p < 0.01 compared to the Con group; ##p < 0. 01 vs. the MI group. b Representative photos of P2X7 immunoreactivity in SG were measured using immunohistochemistry. Histogram shows the IOD of the results. Arrows indicate immunostained neurons. , n = 10. **p < 0.01 compared to the Con group; ##p < 0. 01 vs. the MI group. Scale bars at 20 μm. c Representative photos of P2X7 protein immunostaining in SG were measured using Western blots. Histogram shows the IOD of the results. , n = 6. **p < 0.01, compared to the Con group; ##p < 0. 01 vs. the MI group
P2X7 expression in SG neurons was examined using immunohistochemistry. The intensity of P2X7 immunoreactivity in the MI group was obviously increased compared to the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01, F (5,54) = 29.43). No difference was found between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (4,45) = 2.12). No difference was found between the MI and MI + SC si groups (p > 0.05). The selective P2X7 antagonist BBG, NONRATT021972 siRNA, and P2X7 siRNA inhibited the upregulated expression of P2X7 receptors in SG after myocardial ischemic injury (Fig. 3b).
P2X7 protein expression (normalized to β-actin internal controls) was investigated using Western blotting. The IOD in the MI group was obviously increased compared to the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.0, F (5,30) = 47.66). No difference was found between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (4,25) = 1.91). There was also no difference between the MI group and MI + SCsi group (p > 0.05) (Fig. 3c).
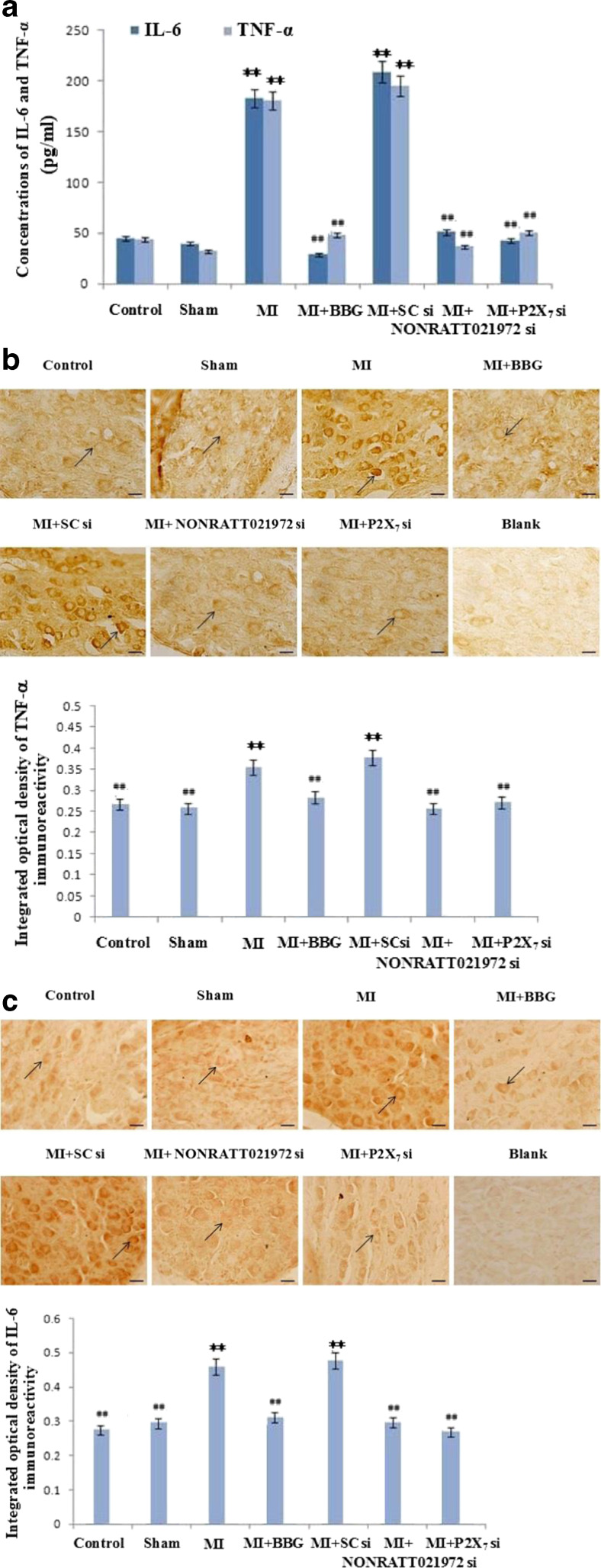
The effects of NONRATT021972 siRNA on serum concentrations of IL-6 or TNF-α, TNF-α expression, or IL-6 immunoreactivity in SG
IL-6 and TNF-α concentrations in blood serum of rats were measured using ELISA. Serum concentration of IL-6 and TNF-α in the MI group was significantly higher than the Con group, Sham group, MI + BBG group, MI + NONRATT021972 si group, and MI + P2X7 si group (p < 0.01, F (IL-6)(5,30) = 6.54, F (TNF-α)(5,30) = 4.72). There were no differences between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (IL-6)(4,25) = 2.27, F (TNF-α)(4,25) = 2.18). There was no difference between the MI group and MI + SC si group (p > 0.05) (Fig. 4a).
Fig. 4.
The concentrations of IL-6 and TNF-α in blood serum and the expression of TNF-α and IL-6 protein immunoreactivity in SG of each group. a Serum concentrations of IL-6 and TNF-α were measured using ELISA. Serum concentrations of IL-6 and TNF-α in the MI group were significantly higher than the Con group, Sham group, MI + BBG group, MI + NONRATT021972 si group, and MI + P2X7 si group (p < 0.01). There were no differences between Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05). There was no difference between the MI group and MI + SC si group (p > 0.05). , n = 6. **p < 0. 01, compared to the Con group; ##p < 0. 01 vs. the MI group. b, c TNF-α expression and IL-6 protein immunoreactivity were detected using immunohistochemistry. Representative photos of TNF-α (b) and IL-6 (c) protein immunoreactivity in SG. Histogram shows the IOD of TNF-α (b) and IL-6 (c) protein immunoreactivity in SG. The expression levels of TNF-α (b) and IL-6 (c) immunoreactivity in the MI group were significantly higher than the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01). No significant differences were observed between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05). Arrows indicate the immunostained neurons. , n = 10. **p < 0.01, compared to the Con group; ##p < 0. 01 vs. the MI group. Scale bars at 20 μm
TNF-α expression was detected using immunohistochemistry. IOD was used to quantify the relative expression levels. TNF-α immunoreactivity was significantly higher in the MI group than the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01, F (5,54) = 37.36). No significant differences were observed between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (4,45) = 1.94) (Fig. 4b).
IL-6 expression in SG was detected using immunohistochemistry. IL-6 immunoreactivity was significantly higher in the MI group than the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01, F (5,54) = 43.57). No significant differences were observed between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (4,45) = 2.76) (Fig. 4c).
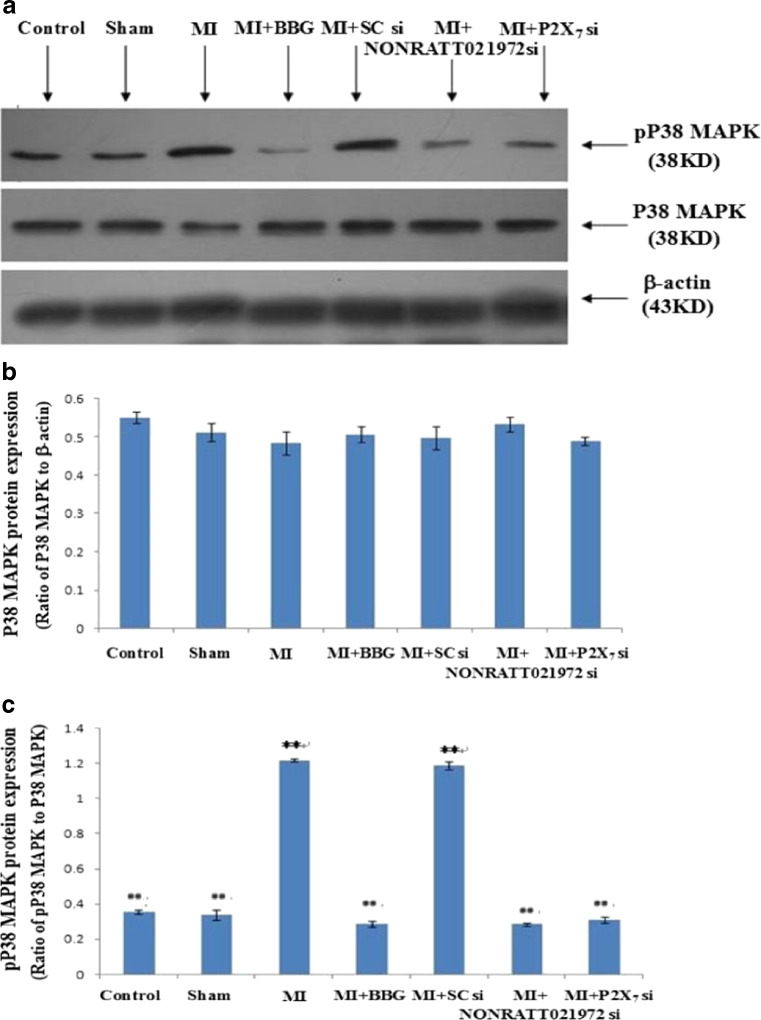
The effects of NONRATT021972 siRNA on p38 MAPK and p-P38 MAPK expression in SG using Western blot
The levels of P38 MAPK and p-P38 MAPK in the SG were analyzed by western blotting. IOD ratio of P38 MAPK to β-actin was not significantly different between the MI group and control group (p > 0.05) (Fig. 5b). However, the IOD ratio of p-P38 MAPK to P38 MAPK (n = 8) in MI group was higher than that in the control group (p < 0.05, n = 8 for each group) (Fig. 5a, c). The results indicated that the role of P38 MAPK phosphorylation in the SG was related to the P2X7 receptor-mediated myocardial ischemic injury in MI rats.
Fig. 5.
The expression of p38 MAPK and p-P38 MAPK protein in SG of each group using Western blots. a Representative photos of p38 MAPK and p-P38 MAPK protein expression in SG. b Histogram shows the ratio of P38 MAPK to β-actin. c Histogram shows the ratio of p38 MAPK to P38 MAPK. The IOD of p-P38 MAPK expression in the MI group was obviously increased compared to the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01). No difference was found between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05). There was also no difference between the MI group and M + SC si group (p > 0.05). , n = 6. **p < 0.01, compared to the Con group; ##p < 0.01 vs. the MI group
In addition, we examined whether the administration of NONRATT021972 siRNA could affect the phosphorylation of P38 MAPK in the MI SG. The IOD ratio of p-P38 MAPK to P38 MAPK in the MI rats treated with NONRATT021972 siRNA in the MI group was obviously increased compared to the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p < 0.01, F (5,30) = 53.43, n = 8 for each group) (Fig. 5a, c). No difference was found between the Con, Sham, MI + BBG, MI + NONRATT021972 si, and MI + P2X7 si groups (p > 0.05, F (4,25) = 2.45). There were no differences in the IOD ratio of P38 MAPK to β-actin between the MI group and MI + scramble siRNA group (p > 0.05, n = 8 for each group) (Fig. 5a, b). Taken together, these results suggest that NONRATT021972 siRNA treatment may contribute to decreasing the phosphorylation and activation of P38 MAPK in the SG of MI rats to alleviate P2X7 receptor-mediated myocardial ischemic injury.
Discussion
Cervical sympathetic ganglia innervate cardiac tissues and regulate myocardial function [39–42]. The altered expression of lncRNAs is involved in the onset and progression of many diseases [7, 9, 43]. Genetic knockouts of some lncRNAs caused developmental defects in mice [44]. Our in situ hybridization experiment demonstrated NONRATT021972 expression in SG. NONRATT021972 expression in SG was significantly higher in the MI group than the control group. Serum NONRATT021972concentration levels were also higher in patients with coronary heart disease than control healthy subjects. This data suggest that the upregulation of NONRATT021972 in SG was related to the pathological process of the myocardial ischemia.
Ligation of the left anterior descending coronary artery causes myocardial hypoxia and induces myocardial ischemic injury. Myocardial ischemic damage increases myocardial cell permeability, which results in elevated myocardial enzymes in peripheral blood [18, 23, 37]. Myocardial enzymes, such as CK-MB, CK, LDH, and AST, are myocardial injury markers [18, 23, 37]. HE staining in this study revealed heart lesions after MI. Serum CK-MB, CK, LDH, and AST concentrations increased in the MI group compared to the control and sham groups. MI rats treated with NONRATT021972 siRNA, the P2X7 antagonist BBG, or P2X7 siRNA exhibited improved histological injury of ischemic cardiac tissues compared MI rats. NONRATT021972 siRNA treatment in MI rats significantly inhibited the ischemic injury-induced elevations in serum CK-MB, CK, LDH, and AST concentrations. Suppression of lncRNA function in vivo attains therapeutic aims [44, 45]. Therefore, NONRATT021972 siRNA treatment may relieve myocardial ischemic injury.
Myocardial ischemia-induced increase in ATP release acts on P2X receptors, which aggravates the myocardial ischemic injury [18, 20, 23, 27, 30, 38]. P2X7 receptors are involved in nervous system function and diseases [11, 16, 17, 46]. P2X7 receptors are expressed in cervical sympathetic ganglia [17, 19, 23, 27]. Our results demonstrated that P2X7 immunoreactivity, mRNA, and protein expression was much higher in MI rats than the control and sham groups. Treatment of MI rats with NONRATT021972 siRNA, BBG, or P2X7 siRNA reduced the expression levels of P2X7 immunoreactivity, mRNA, and protein compared to their myocardial ischemic counterparts. These results suggest that NONRATT021972 siRNA treatment acts on P2X7 receptors to ameliorate myocardial ischemic injury.
Inflammatory mediators augment the nociceptive responses stimulated by ATP [11, 47], and inflammation plays a central role in acute coronary syndrome [18, 48, 49]. Inflammation is the major pathogenetic mechanism associated with myocardial ischemic damage [18, 23, 48–50]. The pro-inflammatory cytokine TNF-α and IL-6 are involved in the pathogenesis of heart ischemic injury [23, 51, 52]. The activation of P2X7 receptors increases the release of inflammatory mediators and cytokines [53, 54]. IL-6 and TNF-α were significantly increased in the MI rats compared to control and sham rats. NONRATT021972 siRNA, BBG, or P2X7 siRNA significantly decreased the enhanced expression of IL-6 and TNF-α after MI injury. The pathogenesis of myocardial ischemia is associated with the balance of anti- and pro-inflammatory cytokines [48, 50]. P2X7 receptors play a significant role in inflammatory diseases [11, 16, 17, 46, 47]. Pharmacological inhibition of P2X7 receptors attenuates inflammatory diseases [11, 46, 47, 55, 56]. NONRATT021972 siRNA may inhibit P2X7 receptor activation to relieve myocardial ischemic inflammatory injury.
MAPKs, such as extracellular signal-regulated kinase (ERK), p38 MAPK, and c-Jun N-terminal kinase (JNK), are serine/threonine kinases [36, 57–59]. Activation of P2X7 receptors activates p38 MAPK [36, 57–59]. Phosphorylation of p38MAP kinase causes activation [36, 57–60]. P2X7 receptor blocker decreased the ATP-provoked activation of p38 MAPK [36, 57, 59]. NONRATT021972 siRNA, BBG, or P2X7 siRNA administration in MI rats in the present study decreased phosphorylated p38 MAPK in SG and improved myocardial ischemic injury. Our data suggest that the improvement of myocardial ischemic injury by P2X7 receptor inhibition was ascribed to the reduction of the p38 MAPK-related signaling pathway. Therefore, gene silencing of NONRATT021972 should inhibit P2X7 activation, reduce p38 MAPK phosphorylation, and prevent the pathological processes of myocardial ischemic injury.
In conclusion, NONRATT021972 expression in SG was significantly higher in the MI group than in the control group. Treatment of MI rats with NONRATT021972 siRNA, BBG, or P2X7 siRNA improved the histological injuries of ischemic cardiac tissue and decreased the elevated serum myocardial enzyme concentrations compared to MI rats. The MI rats treated with NONRATT021972 siRNA, BBG, or P2X7 siRNA reduced the expression of P2X7 immunoreactivity, mRNA, protein, TNF-α, IL-6, and phosphorylated p38 MAPK compared to their myocardial ischemic counterparts. Therefore, NONRATT021972 siRNA treatment inhibited the upregulation of P2X7 receptors in the SG and prevented the pathophysiologic processes mediated by P2X7 receptors in the SG after myocardial ischemic injury.
Acknowledgments
These studies were supported by grants (Nos. 81570735, 31560276, 81560219, 81560529, 81460200, 81171184, 31060139, and 81200853) from the National Natural Science Foundation of China, a grant (No. 20151122040105) from the Technology Pedestal and Society Development Project of Jiangxi Province, a grant (No. 20142BAB205028) from the Natural Science Foundation of Jiangxi Province, and grants (Nos. GJJ13155 and GJJ14319) from the Educational Department of Jiangxi Province.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Lifang Zou and Guihua Tu Joint first authors
Change history
7/8/2020
In the original article, Figs.��2, 3b, and 4b were published incorrectly.
References
- 1.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Liu N, Wang JP, Wang YQ, Yu XL, Wang ZB, Cheng XC, Zou Q. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012;68(4):611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30(8):348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790(9):936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnstock G. Purinergic signalling: from discovery to current developments. Exp Physiol. 2014;99(1):16–34. doi: 10.1113/expphysiol.2013.071951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnstock G, Krugel U, Abbracchio MP, Illes P. Purinergic signalling: from normal behaviour to pathological brain function. Prog Neurobiol. 2011;95(2):229–274. doi: 10.1016/j.pneurobio.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87(2):659–797. doi: 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- 13.Mutafova-Yambolieva VN, Durnin L. The purinergic neurotransmitter revisited: a single substance or multiple players? Pharmacol Ther. 2014;144(2):162–191. doi: 10.1016/j.pharmthera.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erlinge D, Burnstock G. P2 receptors in cardiovascular regulation and disease. Purinergic Signal. 2008;4(1):1–20. doi: 10.1007/s11302-007-9078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassort G. Adenosine 5′-triphosphate: a P2-purinergic agonist in the myocardium. Physiol Rev. 2001;81(2):767–806. doi: 10.1152/physrev.2001.81.2.767. [DOI] [PubMed] [Google Scholar]
- 16.Skaper SD, Debetto P, Giusti P. The P2X7 purinergic receptor: from physiology to neurological disorders. FASEB J. 2010;24(2):337–345. doi: 10.1096/fj.09-138883. [DOI] [PubMed] [Google Scholar]
- 17.Sperlágh B, Vizi ES, Wirkner K, Illes P. P2X 7 receptors in the nervous system. Prog Neurobiol. 2006;78(6):327–346. doi: 10.1016/j.pneurobio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Liu S, Xu B, Li G, Li G, Huang A, Wu B, Peng L, Song M, Xie Q, Lin W, Xie W, Wen S, Zhang Z, Xu X, Liang S. Study of baicalin on sympathoexcitation induced by myocardial ischemia via P2X 3 receptor in superior cervical ganglia. Auton Neurosci. 2015;189:8–15. doi: 10.1016/j.autneu.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Kong F, Liu S, Xu C, Liu J, Li G, Li G, Gao Y, Lin H, Tu G, Peng H, Qiu S, Fan B, Zhu Q, Yu S, Zheng C, Liang S. Electrophysiological studies of upregulated P2X7 receptors in rat superior cervical ganglia after myocardial ischemic injury. Neurochem Int. 2013;63(3):230–237. doi: 10.1016/j.neuint.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Liu S, Yang Y, Xie J, Liu J, Kong F, Tu G, Wu R, Li G, Liang S. Effects of oxymatrine on sympathoexcitatory reflex induced by myocardial ischemic signaling mediated by P2X(3) receptors in rat SCG and DRG. Brain Res Bull. 2011;84(6):419–424. doi: 10.1016/j.brainresbull.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Liu S, Zhang J, Yu K, Xu C, Lin J, Li X, Liang S. Increased sympathoexcitatory reflex induced by myocardial ischemic nociceptive signaling via P2X2/3 receptor in rat superior cervical ganglia. Neurochem Int. 2010;56(8):984–990. doi: 10.1016/j.neuint.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Liang S, Xu C, Li G, Gao Y. P2X receptors and modulation of pain transmission: focus on effects of drugs and compounds used in traditional Chinese medicine. Neurochem Int. 2010;57(7):705–712. doi: 10.1016/j.neuint.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Li G, Peng H, Tu G, Kong F, Liu S, Gao Y, Xu H, Qiu S, Fan B, Zhu Q, Yu S, Zheng C, Wu B, Peng L, Song M, Wu Q, Li G, Liang S. Sensory-sympathetic coupling in superior cervical ganglia after myocardial ischemic injury facilitates sympathoexcitatory action via P2X7 receptor. Purinergic Signal. 2013;9(3):463–479. doi: 10.1007/s11302-013-9367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Yu S, Xu C, Peng L, Xu H, Zhang C, Li G, Gao Y, Fan B, Zhu Q, Zheng C, Wu B, Song M, Wu Q, Liang S. Puerarin alleviates aggravated sympathoexcitatory response induced by myocardial ischemia via regulating P2X3 receptor in rat superior cervical ganglia. Neurochem Int. 2014;70:39–49. doi: 10.1016/j.neuint.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Zhang C, Shi Q, Li G, Song M, Gao Y, Xu C, Xu H, Fan B, Yu S, Zheng C, Zhu Q, Wu B, Peng L, Xiong H, Wu Q, Liang S. Puerarin blocks the signaling transmission mediated by P2X3 in SG and DRG to relieve myocardial ischemic damage. Brain Res Bull. 2014;101:57–63. doi: 10.1016/j.brainresbull.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Shao LJ, Liang SD, Li GL, Xu CS, Zhang CP. Exploration of P2X3 in the rat stellate ganglia after myocardial ischemia. Acta Histochem. 2007;109(4):330–337. doi: 10.1016/j.acthis.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Tu G, Li G, Peng H, Hu J, Liu J, Kong F, Liu S, Gao Y, Xu C, Xu X, Qiu S, Fan B, Zhu Q, Yu S, Zheng C, Wu B, Peng L, Song M, Wu Q, Liang S. P2X(7) inhibition in stellate ganglia prevents the increased sympathoexcitatory reflex via sensory-sympathetic coupling induced by myocardial ischemic injury. Brain Res Bull. 2013;96:71–85. doi: 10.1016/j.brainresbull.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhang CP, Xu CS, Liang SD, Li GL, Gao Y, Wang YX, Zhang AX, Wan F. The involvement of P2X3 receptors of rat sympathetic ganglia in cardiac nociceptive transmission. J Physiol Biochem. 2007;63(3):249–257. doi: 10.1007/BF03165788. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Li G, Liang S, Xu C, Zhu G, Wang Y, Zhang A, Wan F. Myocardial ischemic nociceptive signaling mediated by P2X3 receptor in rat stellate ganglion neurons. Brain Res Bull. 2008;75(1):77–82. doi: 10.1016/j.brainresbull.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Fu LW, Longhurst JC. A new function for ATP: activating cardiac sympathetic afferents during myocardial ischemia. Am J Physiol Heart Circ Physiol. 2010;299(6):H1762–H1771. doi: 10.1152/ajpheart.00822.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasileiou E, Montero RM, Turner CM, Vergoulas G. P2X7 receptor at the heart of disease. Hippokratia. 2010;14(3):155. [PMC free article] [PubMed] [Google Scholar]
- 32.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbeloa J, Perez-Samartin A, Gottlieb M, Matute C. P2X7 receptor blockade prevents ATP excitotoxicity in neurons and reduces brain damage after ischemia. Neurobiol Dis. 2012;45(3):954–961. doi: 10.1016/j.nbd.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, Xu C, Liang S, Zhang A, Mu S, Wang Y, Wan F. Effect of tetramethylpyrazine on primary afferent transmission mediated by P2X3 receptor in neuropathic pain states. Brain Res Bull. 2008;77(1):27–32. doi: 10.1016/j.brainresbull.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 36.Xu H, Wu B, Jiang F, Xiong S, Zhang B, Li G, Liu S, Gao Y, Xu C, Tu G, Peng H, Liang S, Xiong H. High fatty acids modulate P2X(7) expression and IL-6 release via the p38 MAPK pathway in PC12 cells. Brain Res Bull. 2013;94:63–70. doi: 10.1016/j.brainresbull.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Li G, Yu K, Liang S, Wan F, Xu C, Gao Y, Liu S, Lin J. Expressions of P2X2 and P2X3 receptors in rat nodose neurons after myocardial ischemia injury. Auton Neurosci. 2009;145(1-2):71–75. doi: 10.1016/j.autneu.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Li G, Liang S, Zhang A, Xu C, Gao Y, Zhang C, Wan F. Role of P2X3 receptor in myocardial ischemia injury and nociceptive sensory transmission. Auton Neurosci. 2008;139(1-2):30–37. doi: 10.1016/j.autneu.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Armour JA. Potential clinical relevance of the ‘little brain’ on the mammalian heart. Exp Physiol. 2008;93(2):165–176. doi: 10.1113/expphysiol.2007.041178. [DOI] [PubMed] [Google Scholar]
- 40.Boehm S, Kubista H. Fine tuning of sympathetic transmitter release via ionotropic and metabotropic presynaptic receptors. Pharmacol Rev. 2002;54(1):43–99. doi: 10.1124/pr.54.1.43. [DOI] [PubMed] [Google Scholar]
- 41.Gibbins I. Functional organization of autonomic neural pathways. Organogenesis. 2013;9(3):169–175. doi: 10.4161/org.25126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasan W. Autonomic cardiac innervation: development and adult plasticity. Organogenesis. 2013;9(3):176–193. doi: 10.4161/org.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5(22):10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e1749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24(10):594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta N, Kaur M, Singh M, Chand S, Vyas B, Silakari P, Bahia MS, Silakari O. Purinergic receptor P2X 7: a novel target for anti-inflammatory therapy. Bioorg Med Chem. 2014;22(1):54–88. doi: 10.1016/j.bmc.2013.10.054. [DOI] [PubMed] [Google Scholar]
- 47.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112(2):358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Biswas S, Ghoshal PK, Mandal N. Synergistic effect of anti and pro-inflammatory cytokine genes and their promoter polymorphism with ST-elevation of myocardial infarction. Gene. 2014;544(2):145–151. doi: 10.1016/j.gene.2014.04.065. [DOI] [PubMed] [Google Scholar]
- 49.Heo JM, Park JH, Kim JH, You SH, Kim JS, Ahn C, Hong SJ, Shin K, Lim D. Comparison of inflammatory markers between diabetic and nondiabetic ST segment elevation myocardial infarction. J Cardiol. 2012;60(3):204–209. doi: 10.1016/j.jjcc.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Biswas S, Ghoshal PK, Mandal SC, Mandal N. Relation of anti- to pro-inflammatory cytokine ratios with acute myocardial infarction. Korean J Intern Med. 2010;25(1):44–50. doi: 10.3904/kjim.2010.25.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldman AM, Combes A, Wagner D, Kadakomi T, Kubota T, Li YY, McTiernan C. The role of tumor necrosis factor in the pathophysiology of heart failure. J Am Coll Cardiol. 2000;35(3):537–544. doi: 10.1016/S0735-1097(99)00600-2. [DOI] [PubMed] [Google Scholar]
- 52.Matsumori A. Roles of cytokines in the pathogenesis of heart failure. Nihon Rinsho Jpn J Clin Med. 2003;61(5):745–750. [PubMed] [Google Scholar]
- 53.Colomar A, Marty V, Medina C, Combe C, Parnet P, Amedee T. Maturation and release of interleukin-1beta by lipopolysaccharide-primed mouse Schwann cells require the stimulation of P2X7 receptors. J Biol Chem. 2003;278(33):30732–30740. doi: 10.1074/jbc.M304534200. [DOI] [PubMed] [Google Scholar]
- 54.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, Panther E, Di Virgilio F. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176(7):3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 55.Arulkumaran N, Unwin RJ, Tam FW. A potential therapeutic role for P2X7 receptor (P2X7R) antagonists in the treatment of inflammatory diseases. Expert Opin Investig Drugs. 2011;20(7):897–915. doi: 10.1517/13543784.2011.578068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caseley EA, Muench SP, Baldwin SA, Simmons K, Fishwick CW, Jiang LH. Docking of competitive inhibitors to the P2X7 receptor family reveals key differences responsible for changes in response between rat and human. Bioorg Med Chem Lett. 2015;25(16):3164–3167. doi: 10.1016/j.bmcl.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji RR, Suter MR. p38 MAPK, microglial signaling, and neuropathic pain. Mol Pain. 2007;3:33. doi: 10.1186/1744-8069-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S, Ma Q, Krafft PR, Chen Y, Tang J, Zhang J, Zhang JH (2013) P2X7 receptor antagonism inhibits p38 MAPK activation and ameliorates neuronal apoptosis after subarachnoid hemorrhage in the rat. Crit Care Med 41(12):1–9. doi:10.1097/CCM.0b013e31829a8246 [DOI] [PMC free article] [PubMed]
- 59.Xu H, Xiong C, He L, Wu B, Peng L, Cheng Y, Jiang F, Tan L, Tang L, Tu Y, Yang Y, Liu C, Gao Y, Li G, Zhang C, Liu S, Xu C, Wu H, Li G, Liang S. Trans-resveratrol attenuates high fatty acid-induced P2X7 receptor expression and IL-6 release in PC12 cells: possible role of P38 MAPK pathway. Inflammation. 2015;38(1):327–337. doi: 10.1007/s10753-014-0036-6. [DOI] [PubMed] [Google Scholar]
- 60.Papp L, Vizi ES, Sperlagh B. P2X7 receptor mediated phosphorylation of p38MAP kinase in the hippocampus. Biochem Biophys Res Commun. 2007;355(2):568–574. doi: 10.1016/j.bbrc.2007.02.014. [DOI] [PubMed] [Google Scholar]