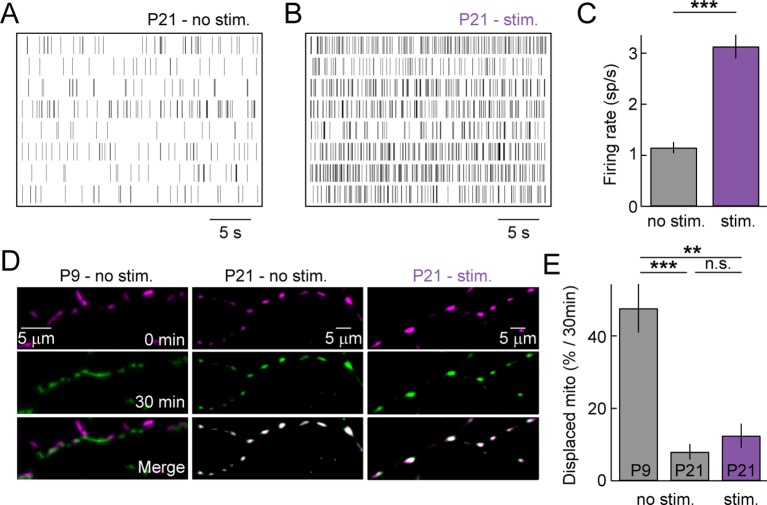
Figure 5. Sensory-evoked neuronal activity and mitochondrial motility at maturity.
(A, B) Spike raster plots of eight representative RGCs recorded in darkness (A) and during presentation of a full-field white noise stimulus (B, see 'Materials and methods'). (C) Bars (error bars) indicate the mean (± SEM) firing rates of RGCs (n = 334 RGCs, 3 retinas, p<10–26). (D) Representative RGCs expressing mtCFP at t = 0 min (top panels), t = 30 min (middle panels) after being exposed to white noise stimulus (right panels) or kept in darkness (left and center panels). Bottom panels show merged images of t = 0 and t = 30 min. (E) Bars (error bars) indicating the mean (± SEM) of % mitochondria displaced between t = 0 min and t = 30 min (P9 – no stim. n = 5 RGCs, P21 – no stim. n = 5 RGCs, P21 – stim n = 5 RGCs). RGCs, retinal ganglion cells.