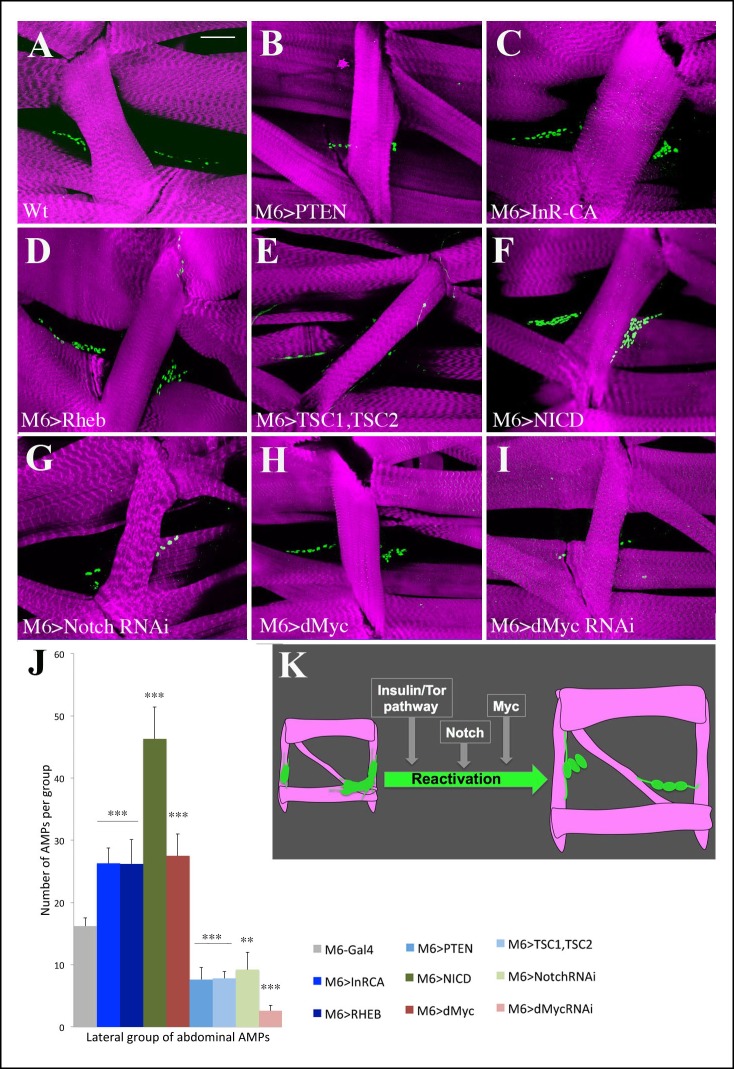
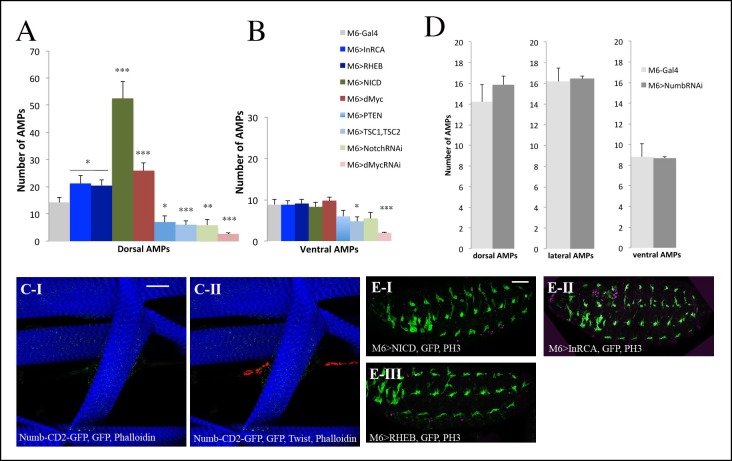
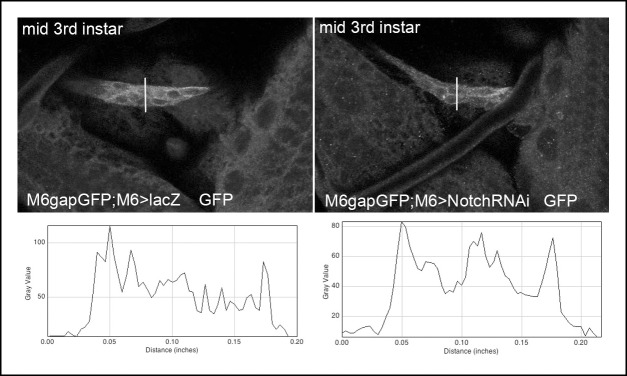
Figure 4. Insulin/TOR and Notch pathways control AMP reactivation in larval stages.
(A–I) Flat preparations of the mid-stage matched third-instar larvae stained for Twist (green) labeling AMP nuclei and stained for Phalloidin (magenta) labeling the larval muscles. The abdominal lateral group of AMPs is shown in (A) representative control larva (M6-Gal4) and (B–I) in larvae with modified Insulin, TOR, Notch and Myc expression. M6-Gal4 driver is used to AMP-specifically drive the expression of: (B) PTEN, an inhibitor of the Insulin pathway; (C) InR-CAAX, a constitutively activated form of insulin receptor; (D) RHEB, an activator of the TOR pathway; (E) TSC1, TSC2, a complex of two proteins that inhibits the TOR pathway; (F) NICD, Notch intracellular domain that constitutively activates the Notch pathway; (G) dsRNA against Notch transcript; (H) overexpression of dMyc; (I) dsRNA against dMyc transcript. (J) Graphical representation of the mean number of lateral AMPs in the different genetic contexts shown in (A–I). (***) indicates P≤0.001. Scale bar: 36 microns. (K) A scheme illustrating the promoter influence of Insulin and Notch pathways and Myc on AMP reactivation.
DOI: http://dx.doi.org/10.7554/eLife.08497.014