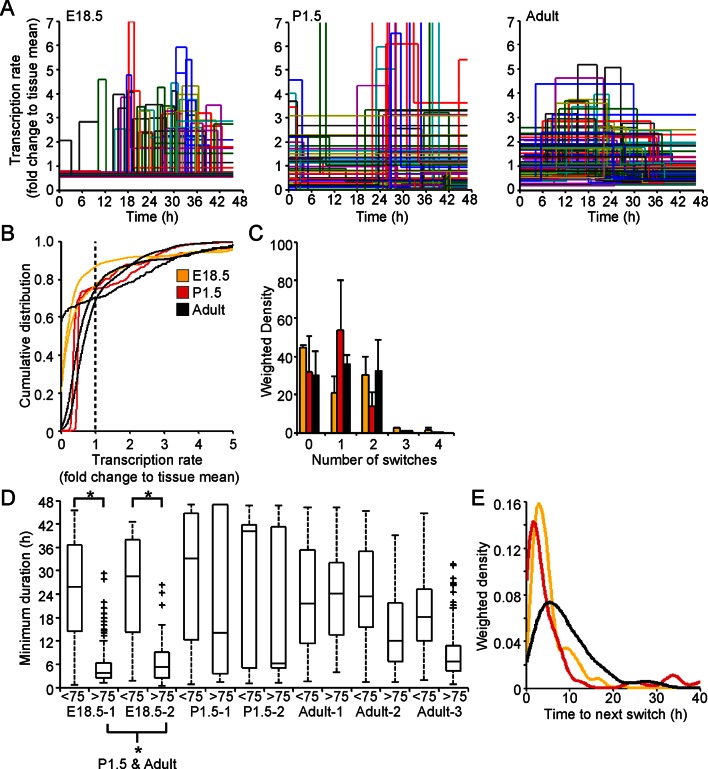
Figure 7. Comparison of prolactin transcription dynamics in different pituitary states.
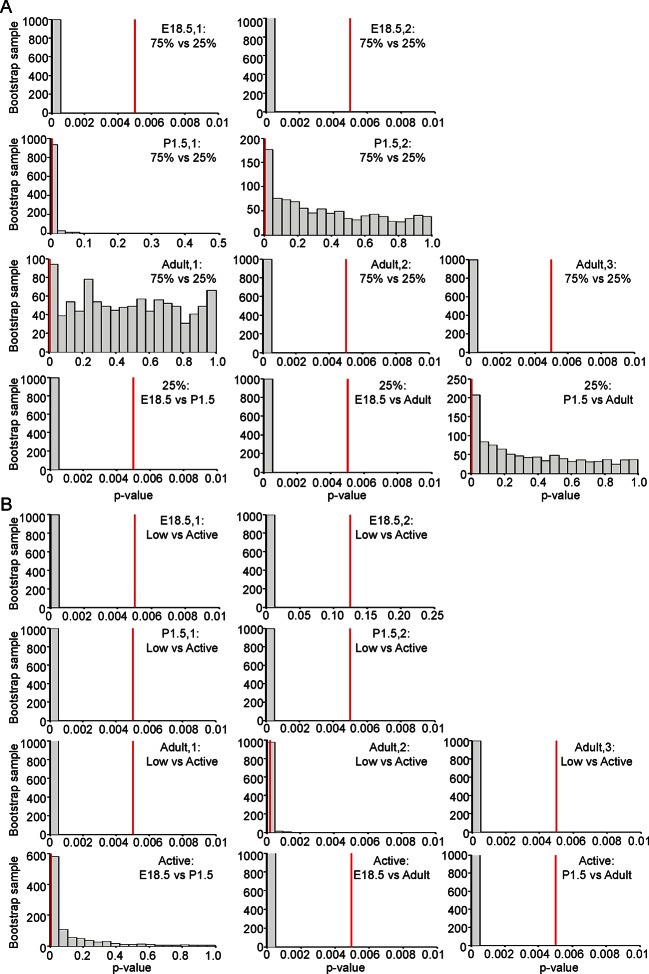
(A) The stochastic switch model enables comparison of transcription dynamics in different pituitary states. Example plots of transcription rates estimated using the stochastic switch model. Analyses of populations of cells are shown from different states of pituitary tissue development. Each cell is associated with a single transcriptional profile obtained by random sampling of all the possible profiles for that cell (shown as in Figure 2A iii, with transcription rate and switch timings depicted). Data shown were from a single experiment representative of independent experiments (i: E18.5 n = 181 cells, ii: P1.5 n = 217 cells, iii: Adult n = 305 cells). (B–E) Characterisation of transcription dynamics estimated using the stochastic switch model. (B) The cumulative distribution function of the frequency of different transcription rates was estimated using a weighted kernel. No clear difference between pituitary developmental states was evident. (C) Weighted histogram of the number of switches for each developmental state. Data, pooled from independent experiments, show the mean + SD, calculated by weighting the number of switches in each profile by the probability of occurrence. P1.5 has significantly fewer switches to Adult and E18.5 (p<0.01 Mann-Whitney U test of the pooled distributions). (D) The duration of transcription rates for each tissue state. Transcription rates were binned into quartiles, with the first three lower quartiles grouped together (<75) as one bin and the highest quartile as the other bin (>75). The duration of transcriptional states was calculated for each bin. The duration represents the minimum duration spent in each transcriptional state, as not all transcriptional states were completely observed within the time-frame of the imaging experiment. Both E18.5 datasets showed a significant difference in the duration spent in low transcriptional states (lower 3 quartiles) to high transcriptional states (upper quartile) as did one P1.5 dataset and two Adult datasets. Moreover, the duration spent in the upper quartile of the E18.5 (pooled data) was significantly different to the duration spent in the upper quartile of either the pooled data of P1.5 or Adult. Significance was assessed through the distribution of the p-values calculated from a Mann-Whitney bootstrap (Figure 7—figure supplement 2). Boxplots were calculated by a random sampling of the weighted transcription rate distributions and the associated durations. (E) The graph shows a kernel density estimate of the time between any two switches using data from panel D, for cells that displayed more than one switch so that the total duration spent in a single transcriptional state was estimated. The inter-switch time in the adult tissues is longer than the inter switch times of either the E18.5 or P1.5 tissues. E18.5 = orange, P1.5 = red, Adult = black. All boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile. SD, standard deviation.
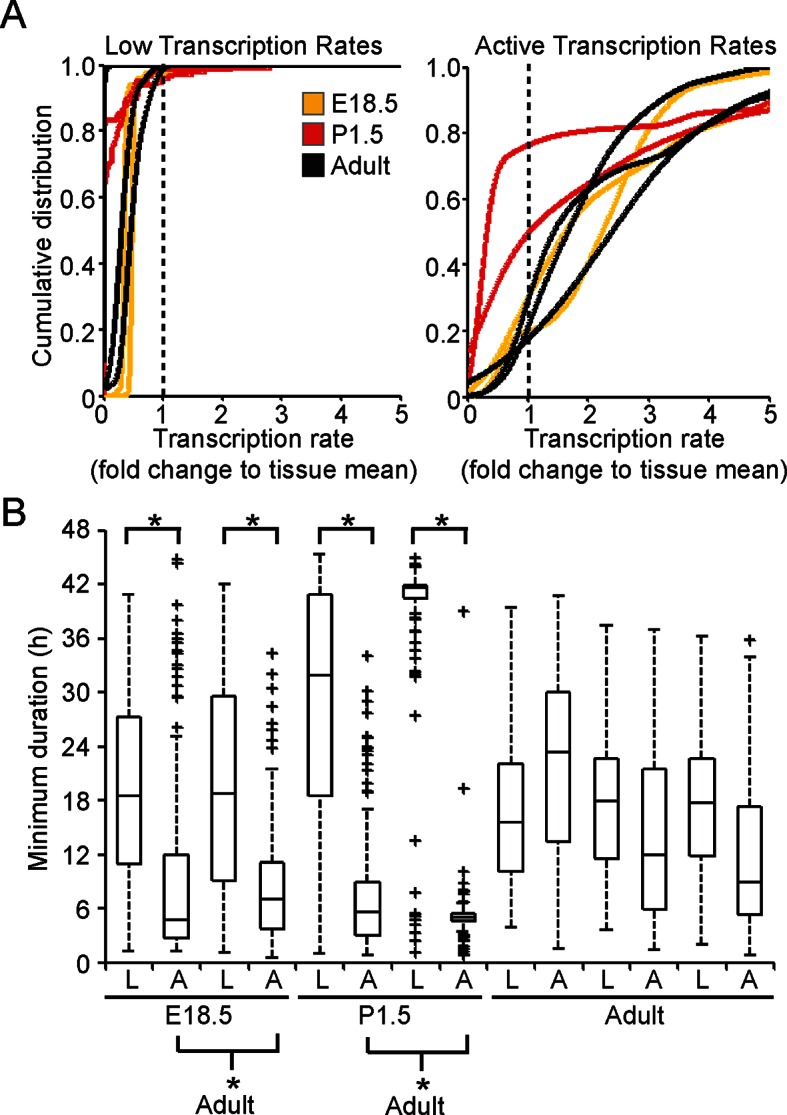
Figure 7—figure supplement 1. Characterisation of prolactin transcription dynamics.