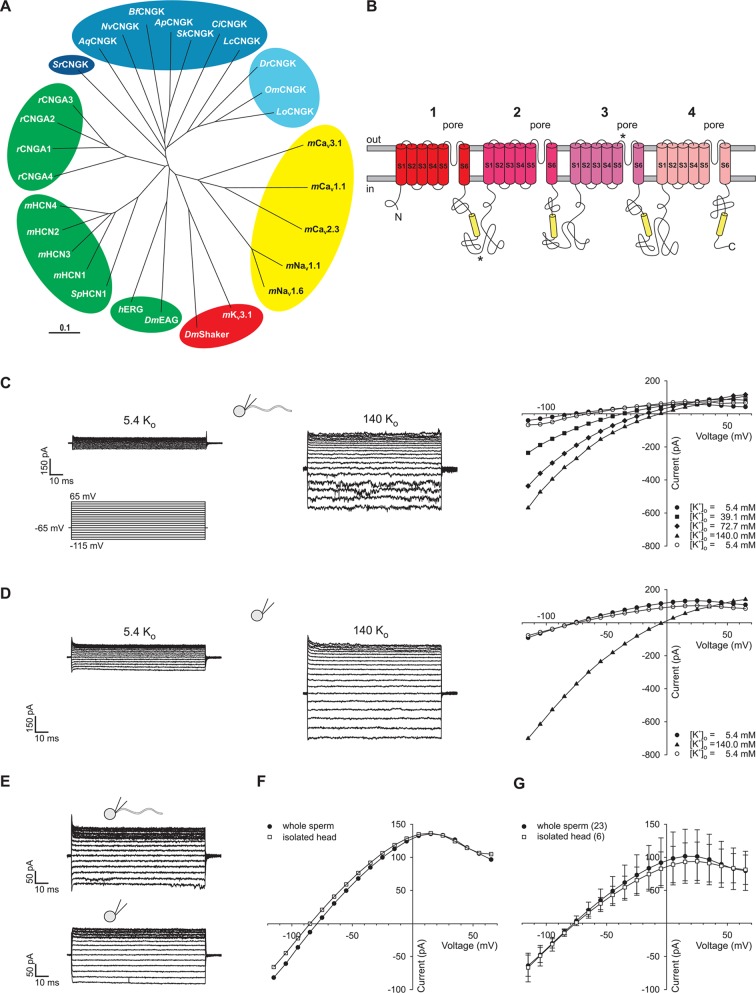
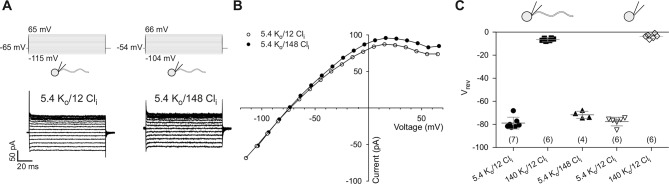
Figure 1. Identification of DrCNGK channel homologues and of a K+ channel in D. rerio sperm.
(A) Phylogenetic tree (Page, 1996) of various ion channel families. The CNGK channel family exists in protozoa (dark blue), marine invertebrates and fish (medium blue), and freshwater fish (light blue). The HCN, CNG, and KCNH channel families are highlighted in green; voltage-gated Nav and Cav channels are highlighted in yellow; and voltage-gated Kv channels are highlighted in red. The following ion channel sequences were used: CNGK channels from zebrafish (DrCNGK), rainbow trout (OmCNGK), spotted gar (LoCNGK), West Indian Ocean coelacanth (LcCNGK), sea urchin (ApCNGK), acorn worm (SkCNGK), amphioxus (BfCNGK), starlet sea anemone (NvCNGK), vasa tunicate (CiCNGK), sponge (AqCNGK), choanoflagellate (SrCNGK); murine HCN channel subunits 1 (mHCN1), 2 (mHCN2), 3 (mHCN3), 4 (mHCN4), and the HCN channel from sea urchin (SpHCN1); rat CNGA subunits A1 (rCNGA1), A2 (rCNGA2), A3 (rCNGA3), and A4 (rCNGA4); the KCNH channels from fruit fly (DmEAG) and human (hERG); murine voltage-gated Nav (mNav 1.1 and mNav 1.6) and Cav channels (mCav1.1, mCav2.3 and mCav3.1) and voltage-gated Kv channels from fruit fly (DmShaker) and mouse (mKv3.1). Full-length Latin names and accession numbers are given in experimental procedures. Scale bar represents 0.1 substitutions per site. (B) Pseudo-tetrameric structure of CNGK channels. Numbers 1 to 4, homologous repeats; S1 to S6, transmembrane segments; yellow cylinders, cyclic nucleotide-binding domain CNBD; asterisks, epitopes recognized by antibodies anti-repeat1 of DrCNGK (polyclonal) and anti-repeat3 of DrCNGK (YENT1E2, monoclonal). (C) Whole-cell recordings from zebrafish sperm at low (left upper panel) and high (middle panel) extracellular K+ concentrations. Left lower panel: Voltage step protocol. Right panel: corresponding IV relations. (D) Whole-cell recordings from an isolated sperm head. Description see part C. (E) Whole-cell recording from zebrafish sperm (upper panel) and an isolated head (lower panel). (F) IV relation of recordings from part E. (G) Pooled IV relations ( ± sd) of currents from zebrafish sperm (filled circle, n = 23) and sperm heads (open squares, n = 6).