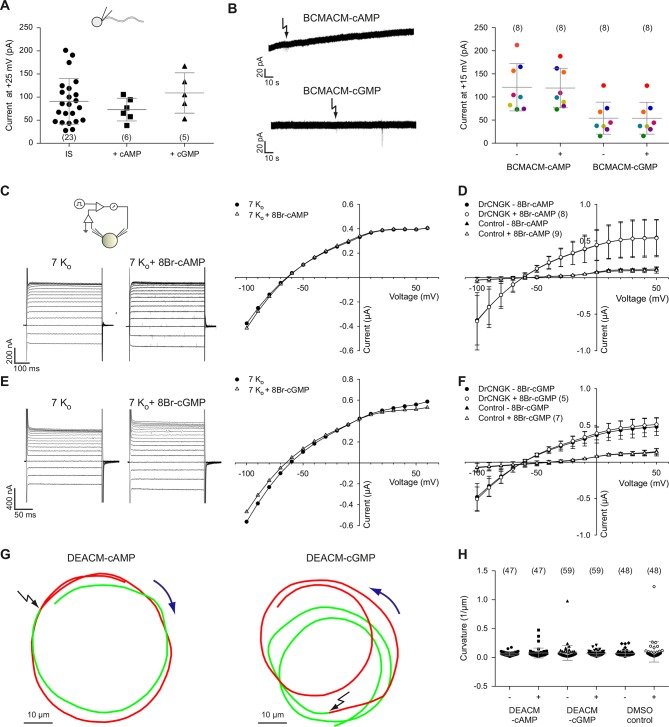
Figure 3. Cyclic nucleotides do not activate K+ channels in sperm.
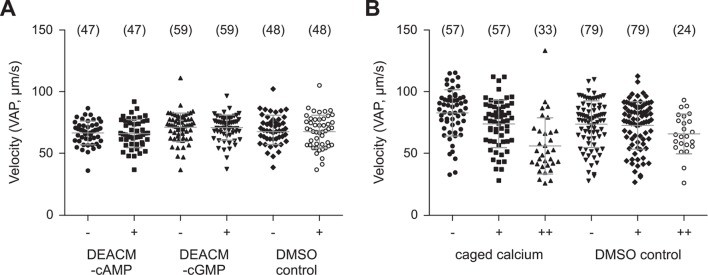
(A) Current amplitude of whole-cell recordings from zebrafish sperm at +25 mV in the absence or presence of 100 µM cAMP or cGMP in the pipette (control: 91 ± 49 pA (n = 23); cAMP: 73 ± 25 pA (n = 6); cGMP: 109 ± 44 pA (n = 5)). Individual data (symbols) and mean ± sd (gray bars), number of experiments in parentheses. (B) Photo-release of cyclic nucleotides from caged precursors inside sperm. Left panel: Whole-cell recordings at +15 mV from sperm loaded with 100 µM BCMACM-caged cAMP (upper panel) or BCMACM-caged cGMP (lower panel). Arrows indicate the delivery of the UV flash to release cyclic nucleotides by photolysis. Right panel: Mean current 3 s before (-) and 3 s after (+) the release of cAMP or cGMP. Statistics as in part A. Data points from individual sperm are indicated by identical colours. (C-F) Currents of heterologously expressed DrCNGK channels in the absence or presence of 8Br- analogs of cyclic nucleotides. (C) Left: Two-Electrode Voltage-Clamp recordings from DrCNGK-injected Xenopus oocytes. Currents shown are in the absence (left traces) and presence (right traces) of 10 mM 8Br-cAMP. Voltage steps as shown in Figure 3—figure supplement 1A. Right: IV relations of current recordings from the left panel. (D) Pooled IV curves from DrCNGK injected and control oocytes; recordings in the absence and presence of 10 mM 8Br-cAMP. (E) Left: Two-Electrode Voltage-Clamp recordings from DrCNGK injected Xenopus oocytes. Currents shown are in the absence (left traces) and presence (right traces) of 10 mM 8Br-cGMP. Right: IV relations of current recordings from the left panel. (F) Pooled IV curves from DrCNGK-injected and control oocytes; recordings in the absence and presence of 10 mM 8Br-cGMP. (G) Swimming path before (green line) and after (red line) photo-release (black flash) of cAMP (left panel) or cGMP (right panel). The blue arrow indicates the swimming direction. Photo-release of cyclic nucleotides was verified by monitoring the increase of fluorescence of the caging group (Figure 3—figure supplement 5) (Hagen et al., 2003). (H) Path curvature before (-) and after (+) release of cAMP or cGMP. Sperm were loaded with 30 µM DEACM-caged cAMP or DEACM-caged cGMP. Statistics as in part A.