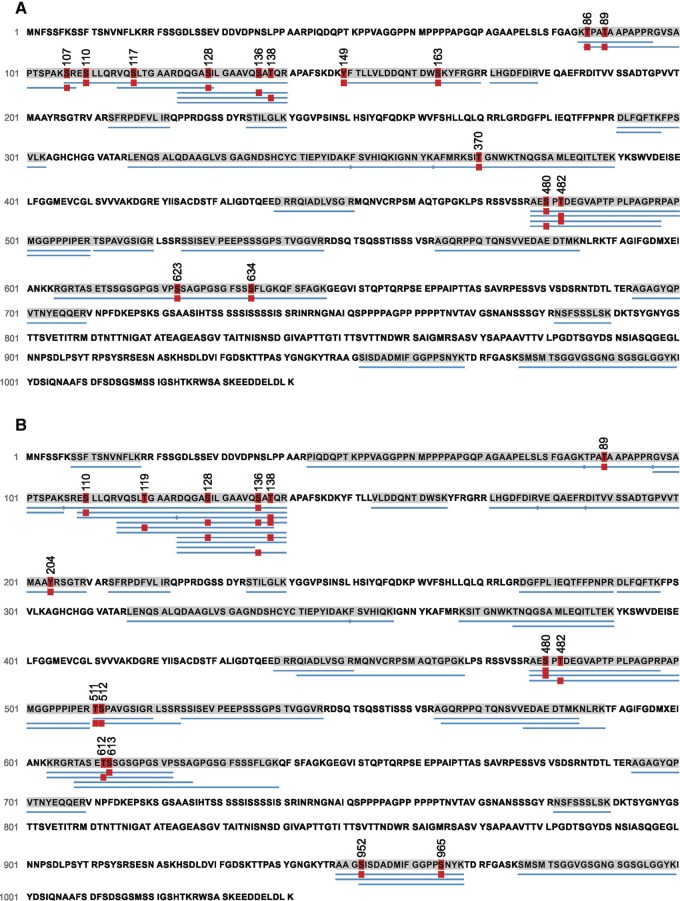
Figure 6.
The pattern of Synapsin phosphorylation is altered in sap47156 mutants. Phosphorylation sites of Synapsin in experimentally naive wild-type WT2 (A) and sap47156 mutant larvae (B). Thirteen LC-MS/MS runs were performed to analyze the phosphorylation status across the Synapsin protein in both genotypes. The number of times a phosphopeptide or its corresponding nonphosphorylated counterpart was detected is indicated as counts in Table 1. (A) We identified 15 phosphosites of Synapsin in the wild-type WT2 and (B) 15 phosphorylated sites of Synapsin in the sap47156 mutant larvae. Blue bars below the sequence indicate the peptides identified as peptide-spectra matches (PSM) using the PEAKS de novo sequencing algorithm. The red “P” boxes indicate phosphorylation (P < 0.005). As an example how to read this display and Table 1, in the wild-type WT2 all peptides covering amino acids 478–497 were found to be phosphorylated at either S480 or S482, but in no case were both or neither of these two found to be phosphorylated. Table 1 then shows that a phosphorylated S480 site was found for 8 out of 15 peptides, while for S482 phosphorylation was observed for the remaining 7 peptides.