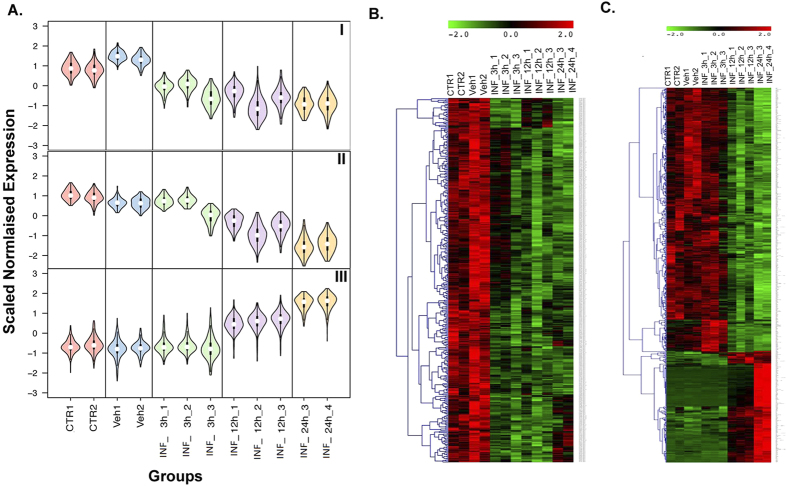
Figure 2. Significant transcriptional changes induced due to infection.
(A) Self-organizing map (SOM) analysis of 1,234 genes commonly differentially expressed among different post-infection time points compared to controls. The SOM maps were clustered into ‘constitutive’ (similar expression changes at 3 h, 12 h, and 24 h) and ‘temporal’ (progressive expression changes at 3 h, 12 h, and 24 h) expression changes on the basis of structure of expression pattern. The figure shows violin plots of selected 3 maps depicting constitutive down-regulation (I), temporal down-regulation (II) and temporal up-regulation (III) patterns. Each cluster represents a set of genes that depict similar expression patterns and are biologically linked to a specific function. The X-axis represents different time points and groups, and the Y-axis represents gene expression on pseudoscale from −3 to +3. (B) Heatmap of constitutive genes depicting similar expression changes at 3 h, 12 h, and 24 h post-infection. (C) Heatmap of temporal genes depicting progressive expression changes from 3 h, 12 h, and 24 h post-infection. Columns represent samples, and rows represent genes. Gene expression levels are shown on a pseudocolor scale (−1 to 1), with red denoting a high expression level and green denoting a low expression level.