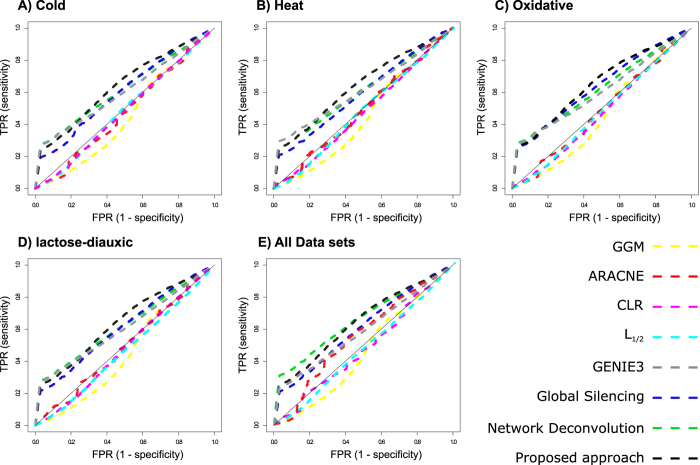
Figure 2. ROC curves for the methods considered in the comparative analysis (E. coli data sets).
ROC curves are shown for the gene regulatory networks predicted based on the data sets from (A) cold, (B) heat, and (C) oxidative stress as well as (D) lactose-diauxic shift in addition to the (E) combination of all four data sets by using the following methods: GGM, ARACNE, CLR, L1/2, GENIE3, Global silencing, Network deconvolution, and the proposed approach based on the fused LASSO which simultaneously considers all four data sets. Due to the high similarity in the performance of the regularization-based models, the ROC curves of L1/2 model is illustrated as the representative to avoid curve overlapping. The color of the dashed lines represents the methods. TPR and FPR stand for true positive rate and false positive rate, respectively.