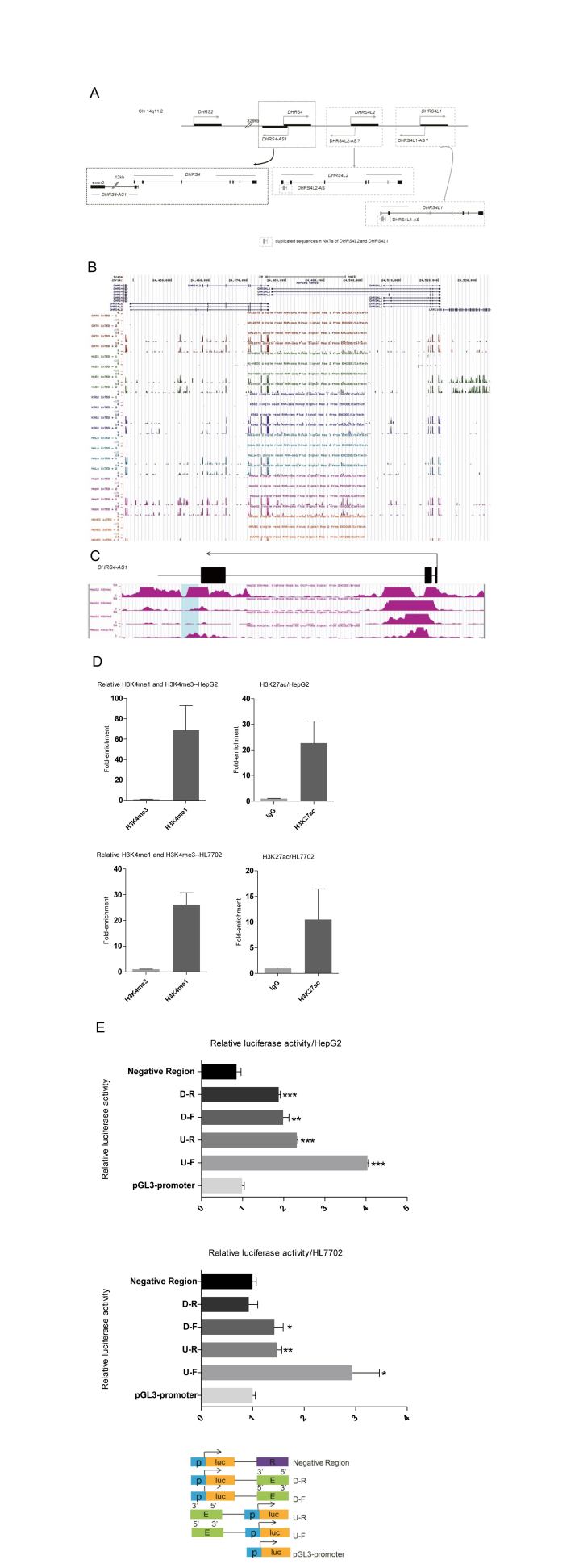
Figure 1. A predicted downstream enhancer region of DHRS4-AS1 shows enhancer activity.
(A) Structure of the human DHRS4 gene cluster and corresponding putative NATs at chromosomal band 14q11.2. In the gene duplication events that gave rise to DHRS4L2 and DHRS4L1 from DHRS4, at least some of the sequences of DHRS4-AS1 present in DHRS4 could, in principle, be retained in DHRS4L2 and DHRS4L1. The gray box (bottom) represents the duplicated sequences of DHRS4-AS1 that were retained in DHRS4L2 and DHRS4L1. (B) ENCODE tracks from the UCSC Genome Browser using strand-specific RNA-seq failed to identify evidence of transcription of DHRS4L2-AS and DHRS4L1-AS from the minus strand in GM12818, H1-hESC, K562, HeLa-S3, HepG2 and HUVEC cells. (C) Localization of the putative enhancer element at the 3′ end of the DHRS4-AS1 gene. ENCODE tracks from the UCSC Genome Browser illustrate the high levels of H3K4me1 and H3K27ac, and the low levels of H3K4me3 in HepG2 cells. The light blue rectangle represents the predicted enhancer region. (D) HepG2 and HL7702 cells were subjected to ChIP for verification of H3K4me1, H3K4me3 and H3K27ac. The abundance of histone modifications within the predicted enhancer region is in accordance with the ENCODE tracks of histone modifications from the UCSC Genome Browser database. (E) The downstream predicted enhancer elevates promoter-driven luciferase activity. The four constructs [upstream/forward (U-F), upstream/reverse (U-R), downstream/forward (D–F) and downstream/reverse (D–R)] were co-transfected with a Renilla reporter gene into HepG2 and HL7702 cells. Random DNA (R) represents the negative control harboring no enhancer features. Luciferase activity was normalized to Renilla luciferase activity and then divided by the values for the pGL3 promoter empty vector control. The positions and orientations of the constructs are shown (lower panel). All data shown are the mean ± standard error of the mean (SEM) of at least three independent experiments. P values were determined by Student’s unpaired two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001.