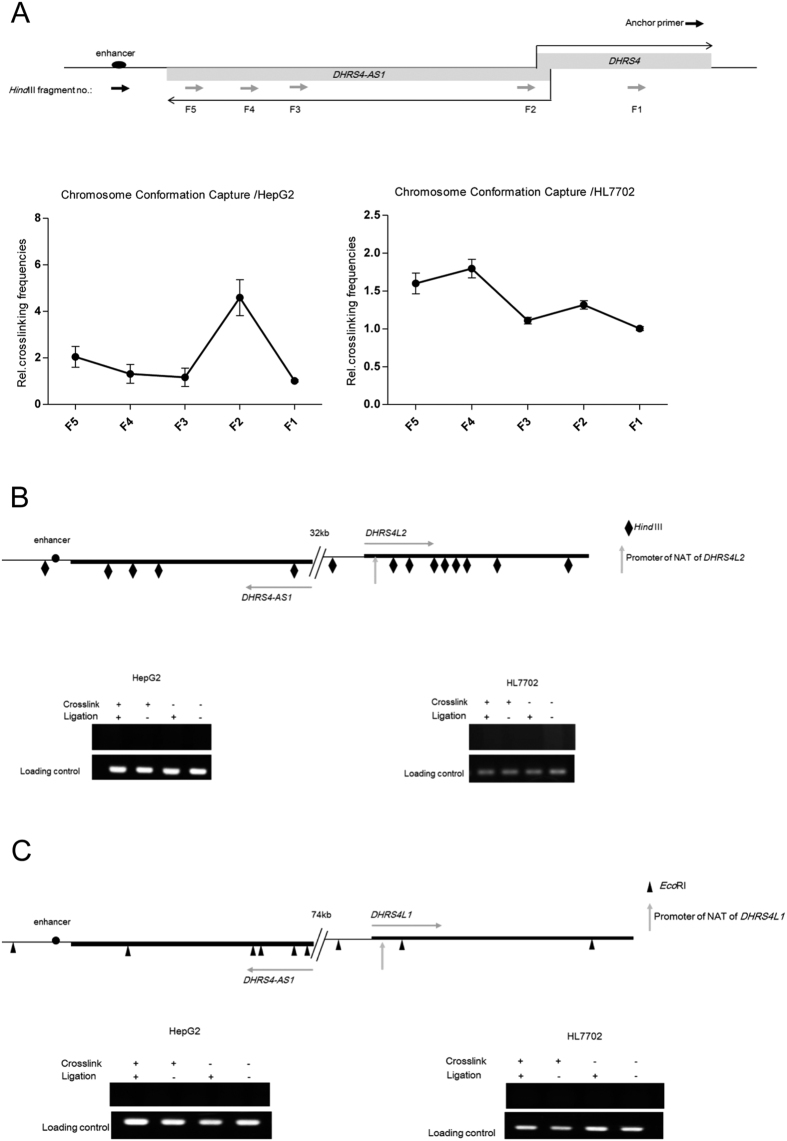
Figure 2. AS1 enhancer selectively interacts with DHRS4-AS1 promoter.
Cells were crosslinked with 1% formaldehyde, and then the reaction was stopped by the addition of glycine. Restriction enzymes HindIII or EcoRI were used to digest the crosslinked chromatin. Then ligation was performed by incubation with T4 DNA ligase. Purified ligation products were determined by qPCR. (A) The relative crosslinking frequencies between the anchor region (the AS1 enhancer) and distal fragments (F1~F5) were measured by qPCR and normalized to the control region (fragment F1). Error bars indicate the mean ± SEM of three experiments. P values were determined by Student’s unpaired two-tailed t test. (B,C) Spatial interactions between the AS1 enhancer and homologous promoter regions of putative DHRS4L2 and DHRS4L1 NATs were determined by 3C array. Crosslinked chromatin was then digested with HindIII (B) or EcoRI (C), followed by ligation. 3C samples from HindIII- (B) or EcoRI- (C) digested crosslinked chromatin without ligation and non-crosslinked genomic DNA with or without ligation were used as negative controls. To ensure the various primer pairs all worked as intended, we performed a random ligation control where PCR was performed to amplify DNA from BAC clones. The primers used here effectively amplified the control templates that contained all the ligation products. Notably, the putative promoter sequences used in this analysis contain all the potential promoters predicted from PROSCAN (see Methods) at the putative homologous NATs of DHRS4L2 and DHRS4L1. The PCR products from 3C samples, not cut with any restriction enzyme, were used as the loading control.