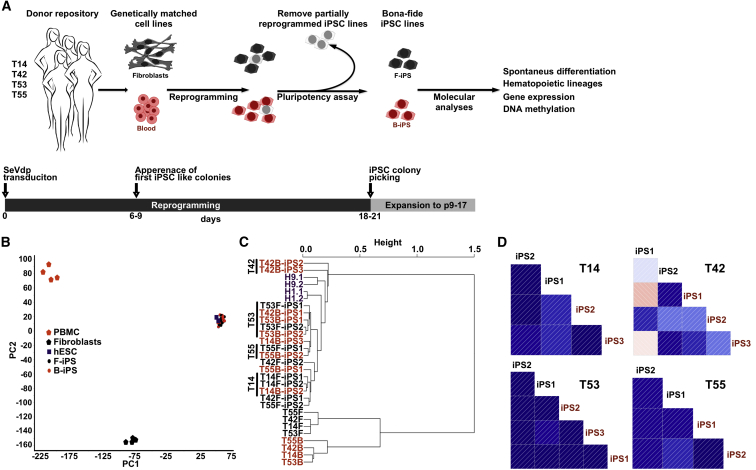
Figure 1.
iPSCs Derived from Fibroblasts and Blood Cells Are Transcriptionally Similar
(A) Schematic representation of the study. Genetically matched iPSC were produced from fibroblasts and blood cells (PBMCs) from four female donors (T14, T42, T53, T55) using Sendai virus (SeVdp) mediated reprogramming. The cell lines used in this study are listed in Table 1. Fibroblast-derived iPSCs (F-iPS) and blood-derived iPSCs (B-iPS) are shown in black and red, respectively, throughout the figures.
(B) PCA of global gene expression data of genetically matched F-iPSCs (n = 8) and B-iPSCs (n = 10) derived from four donors, two human embryonic stem cell lines (hESC), and somatic cells of origin (n = 4/4). Characterization of iPSC lines is presented in Figure S1.
(C) Unsupervised hierarchical clustering of global DNA methylation profiles in genetically matched F-iPSC (n = 8) and B-iPSC (n = 10) lines, somatic cells of origin (n = 4/4), and two hESC lines (H1, H9) performed on a single-nucleotide level using reduced representation bisulfite sequencing.
(D) Pairwise correlation of the genetically matched F- and B-iPSC lines for each donor (T14, n = 4; T42, n = 5; T53, n = 5; T55, n = 4) after local-pooled-error test. The entire list of genes for each donor is presented in Table S1. The direction of the correlation is visualized using thin lines inside boxes, and the magnitude of correlation using the colors. Darker color corresponds to the higher correlation. Isogenic iPSC lines derived from donor T42 display the lowest correlation.