Figure 2.
Methylomic Analyses Demonstrate Minimal Contribution of Source-Cell-Specific Differences to iPSC Variability
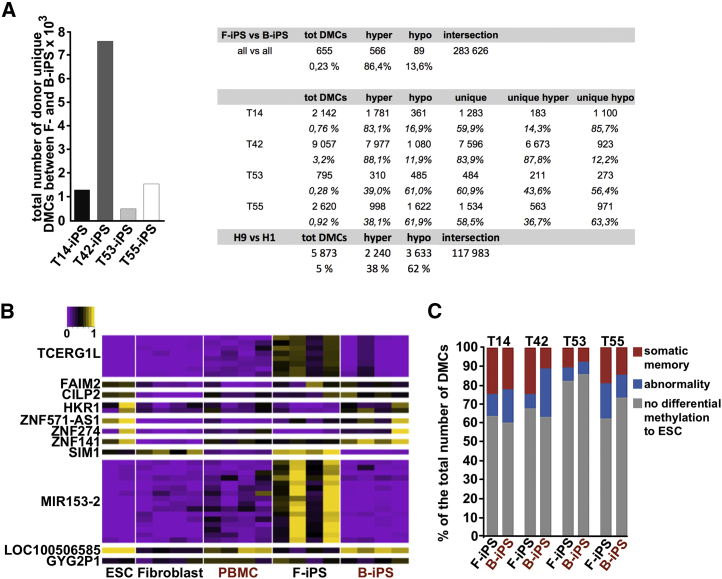
(A) Total number of donor-unique, differentially methylated cytosines (DMCs) between genetically matched iPSC lines derived from fibroblasts and blood cells. Analyses were performed for each donor (T14, n = 4; T42, n = 5; T53, n = 5; T55, n = 4). The table shows the number of DMCs per pairwise comparison as well as the number of hypermethylated CpGs and hypomethylated CpGs in B-iPSCs with respect to F-iPSCs and H1 hESCs with respect to H9. RRBS data for H1 hESCs were obtained from the ENCODE project (Meissner et al., 2008).
(B) A heatmap representing methylation level of the 34 common DMCs across all pluripotent stem cells (n = 20) and somatic cell lines (n = 8). DMCs were annotated at the nearest transcription start site. ESC, embryonic stem cells; PBMC, blood cells; F-iPS, iPSCs derived from fibroblast; B-iPS, iPSCs derived from blood.
(C) Aberrant methylation and somatic cell memory in genetically matched F- and B-iPSC lines (T14, n = 4; T42, n = 5; T53, n = 5; T55, n = 4) compared with embryonic stem cells (ESC). The bar chart represents the percentage of the total number of DMCs. The list of values can be found in (A). Donors are indicated above the bars.