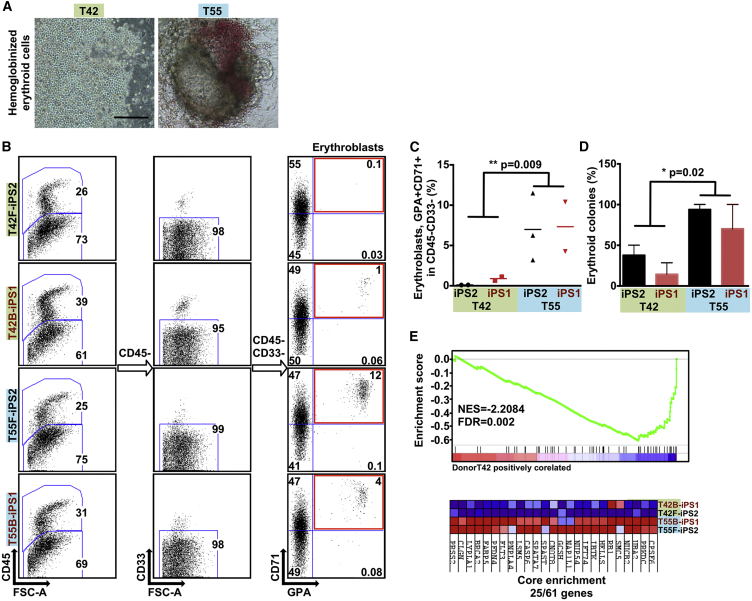
Figure 5.
iPSCs Derived from Different Donors Show Variable Erythroid Lineage Differentiation Propensity
(A) Representative micrographs showing the hematopoietic cells differentiated from iPSCs with reduced hemoglobinized erythroid cells in iPSCs derived from T42 (left panel). Scale bar, 100 μm.
(B) Representative FACS plots of differentiated genetically matched F- and B-iPSC lines (n = 4) showing the percentage of erythroblasts (GPA+ CD71+ gated from CD45− CD33−).
(C) Scatter graphs presenting percentage of erythroblasts generated from the genetically matched F- and B-iPSC lines. Each dot represents an independent experiment.
(D) Percentage of erythroid colonies out of the total colony number generated from iPSC lines. Values are mean ± SEM. Statistics performed by Student's t test, in three or four biological replicates.
(E) Gene set enrichment plot and heatmap showing the genes constituting the core enrichment of overlap between multi-potent progenitors sorted from the bone marrow of DBA patients and iPSC lines derived from T42 cells. T42 iPSC lines show decreased expression levels of genes that are downregulated in Diamond-Blackfan anemia patients.