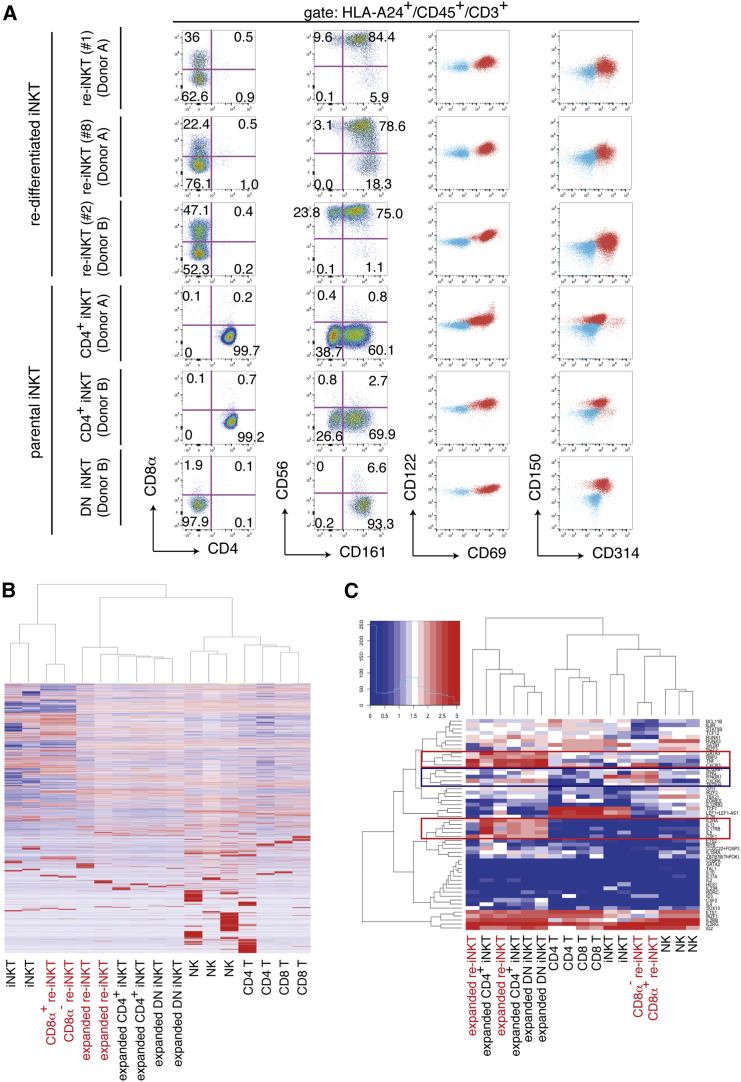
Figure 2.
Gene and Molecular Expression Profile of Re-iNKT Cells
(A) Flow cytometric analysis of T cells re-differentiating from CD4+ iNKT-iPSCs on day 10 after TCR stimulation. iPSC-derived re-differentiating cells were stimulated with α-GalCer-pulsed (10 ng/ml) and irradiated PBMCs from an HLA-A24-negative donor. The results shown are the surface protein expression patterns of re-differentiating cells gated by HLA-A24, CD45, CD3, and 6B11, and are representative of four experiments. The blue and red populations in the right two panels respectively indicate unstained and stained samples for the indicated molecules.
(B and C) Global gene expression profiles (B) and two-way clustering of 53 selected gene expression profiles (C) related to NK/T cell differentiation and function for re-iNKT cells (clones 2 and 4; both from donor B) and primary immune cells. Levels of mRNA expression in re-iNKT cells and primary immune cells were determined using RNA sequencing. Heatmaps show the differential expression of genes among samples. Red and blue indicate increased and decreased expressions, respectively. Blue and red boxes in (C) indicate gene clusters preferentially up-regulated in resting and activated re-iNKT cells, respectively.