Fig. 2.
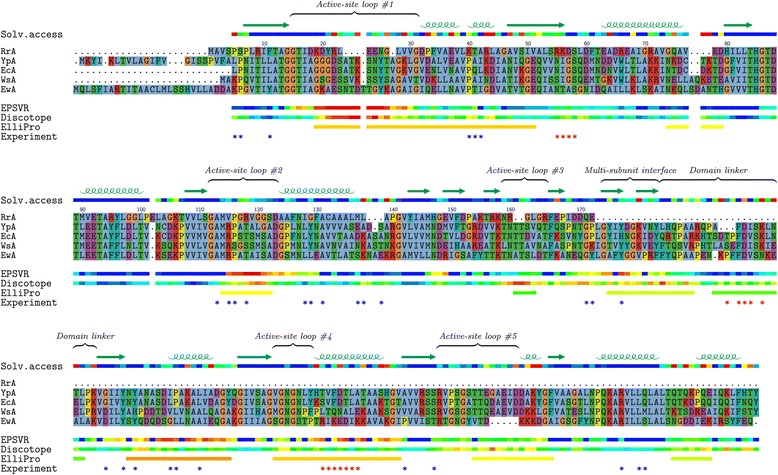
Structure-based multiple sequence alignment of five studied L-asparaginases. Four lines below the alignment show putative epitope region predicted by EPSVR, Discotope, and ElliPro methods, respectively, as well as the experimentally known epitopes projected from the Erwinia chrysanthemi L-asparaginase. Lines above the alignment represent the secondary structure and the estimation of solvent accessibility of EcA tetramer
