Figure 7.
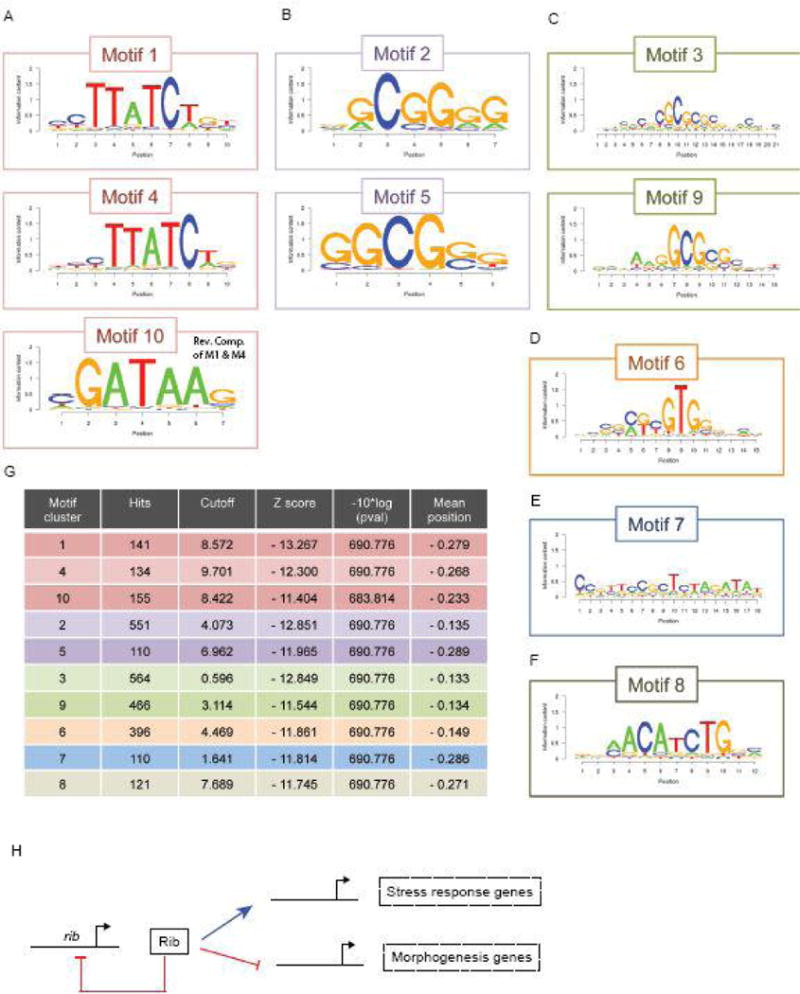
The top ten consensus sites found within Rib bound DNA fragments from the ChIP-seq analysis reveal conserved core motifs. (A) Motif 1 and Motif 4 are displaced by a single nucleotide and Motif 10 is the reverse complement of the most conserved sequence within Motifs 1 and 4. (B and C) Similarly, Motifs 2 and 5 contain a shared core sequence, as do Motifs 3 and 9. (D–lF) Motifs 6 – 8 are distinct, although the N-terminal portion of Motif 6 is related to Motifs 3 and 9. (G) Table of top 10 Motif clusters with the number of hits, the Cutoff, Z score, p value and mean position is shown. (H) A hypothetical model of the Rib gene regulatory network in the context of SG development.
