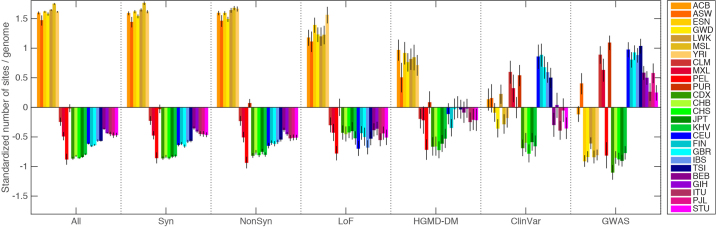
Extended Data Figure 4. The standardized number of variant sites per genome, partitioned by population and variant category.
For each category, z-scores were calculated by subtracting the mean number of sites per genome (calculated across the whole sample), and dividing by the standard deviation. From left: sites with a derived allele, synonymous sites with a derived allele, nonsynonymous sites with a derived allele, sites with a loss-of-function allele, sites with a HGMD disease mutation allele, sites with a ClinVar pathogenic variant, and sites carrying a GWAS risk allele.