Abstract
In clinical trials of coformulated elvitegravir (EVG), cobicistat (COBI), emtricitabine (FTC), and tenofovir disoproxil fumarate (TDF), emergent drug resistance predominantly involved the FTC resistance substitution M184V/I in reverse transcriptase (RT), with or without the tenofovir (TFV) resistance substitution K65R, accompanied by a primary EVG resistance substitution (E92Q, N155H, or Q148R) in integrase (IN). We previously reported that the RT-K65R, RT-M184V, and IN-E92Q substitutions lacked cross-class phenotypic resistance and replicative fitness compensation. As a follow-up, the in vitro characteristics of mutant HIV-1 containing RT-K65R and/or RT-M184V with IN-Q148R or IN-N155H were also evaluated, alone and in combination, for potential interactions. Single mutants displayed reduced susceptibility to their corresponding inhibitor classes, with no cross-class resistance. Viruses with IN-Q148R or IN-N155H exhibited reduced susceptibility to EVG (137- and 40-fold, respectively) that was not affected by the addition of RT-M184V or RT-K65R/M184V. All viruses containing RT-M184V were resistant to FTC (>1,000-fold). Mutants with RT-K65R had reduced susceptibility to TFV (3.3- to 3.6-fold). Without drugs present, the viral fitness of RT and/or IN mutants was diminished relative to that of the wild type in the following genotypic order: wild type > RT-M184V ≥ IN-N155H ≈ IN-Q148R ≥ RT-M184V + IN-N155H ≥ RT-M184V + IN-Q148R ≥ RT-K65R/M184V + IN-Q148R ≈ RT-K65R/M184V + IN-N155H. In the presence of drug concentrations approaching physiologic levels, drug resistance counteracted replication defects, allowing single mutants to outcompete the wild type with one drug present and double mutants to outcompete single mutants with two drugs present. These results suggest that during antiretroviral treatment with multiple drugs, the development of viruses with combinations of resistance substitutions may be favored despite diminished viral fitness.
INTRODUCTION
Evolution of multiple HIV mutations occurs during prolonged antiretroviral (ARV) treatment failure. These mutations typically can be categorized as polymorphic, resulting in no phenotypic change to the virus, or resistance associated, resulting in reduced susceptibility to one or more ARV inhibitors. Additional accessory mutations may also develop to produce combined effects on viral replicative fitness and/or drug susceptibility. Several such relationships between mutations within the same coding region of a target enzyme have been characterized; for example, the G140S substitution in integrase (IN-G140S) has been shown to restore viral fitness of the IN-Q148H substitution and enhance resistance to raltegravir (RAL), an integrase strand transfer inhibitor (INSTI) (1, 2). Additionally, substitutions in one coding region may exhibit effects on susceptibility to ARV inhibitors from a different drug class: for example, the addition of IN-G140S/Q148R to a virus with the nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance substitution K103N appears to significantly enhance resistance to the NNRTI efavirenz (EFV) (3). While data on such cross-class interactions is limited (4, 5), reverse transcriptase (RT) and integrase (IN) are expressed together in the same polyprotein and are proximally associated in replication and preintegration complexes, suggesting possible functional interaction (6–8). Furthermore, INSTI resistance substitutions have been found to develop in viral isolates with existing protease (PR) and RT drug resistance substitutions from treatment-experienced patients (4, 5, 9–11), thus warranting further investigation of possible cross-class interactions in multidrug-resistant HIV.
During the phase 3 clinical trials of a single-tablet regimen consisting of the INSTI elvitegravir (EVG), the pharmacoenhancer cobicistat (COBI), and the nucleoside/nucleotide RT inhibitors (NRTIs) emtricitabine (FTC) and tenofovir disoproxil fumarate (TDF), the majority of HIV-1 isolates with emergent drug resistance contained the FTC resistance substitution M184V/I in RT accompanied by a primary INSTI resistance substitution in IN (T66I, E92Q, T97A, Q148R, or N155H) (12–16). More rarely, the tenofovir (TFV) resistance substitution RT-K65R also developed in addition to RT-M184V/I and the INSTI resistance substitutions IN-E92Q, IN-Q148R, and IN-N155H (12–16). Clonal sequence analysis determined that these INSTI and NRTI resistance substitutions were present together on the same viral genomes (17); however, the commercial assays used to analyze patient isolates during the trials did not amplify RT and IN together in the same assay. Therefore, site-directed mutants representing patterns of resistance found in patient isolates were constructed to evaluate possible cross-class effects on drug susceptibility or viral fitness. We recently reported that the RT-K65R, RT-M184V, and IN-E92Q substitutions altered susceptibility to only their corresponding inhibitor classes, with each substitution cumulatively contributing to decreased viral fitness in the absence of drug pressure (18). Here, we characterize two additional INSTI resistance substitutions, IN-Q148R and IN-N155H, in combination with RT-M184V and RT-K65R.
MATERIALS AND METHODS
Compounds and cells.
The ARV inhibitors EVG, RAL, FTC, TFV, EFV, and darunavir (DRV) were synthesized at Gilead Sciences (Foster City, CA). Zidovudine (AZT) was purchased from Sigma-Aldrich (St. Louis, MO), and dolutegravir (DTG) was purchased from Shanghai Medicilon Inc. (Shanghai, China). MT-2 cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Germantown, MD). HEK 293T cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA).
Site-directed mutant viruses and antiviral drug susceptibility assay.
Chimeric HIV-1 xxLAI proviral plasmids containing NRTI and INSTI resistance mutations were constructed as previously described (18–23). Viruses were generated by transient transfection of HEK 293T cells using TransIT-293 (Mirus Bio LLC, Madison, WI), and titers were determined in triplicate on MT-2 cells by cytopathic effect to determine the 50% tissue culture infectious dose (TCID50) 5 days postinfection (24). Drug susceptibilities of wild-type and mutant viruses to NRTIs, INSTIs, NNRTIs, and protease inhibitors (PIs) were determined in MT-2 cells using a 5-day multiple-cycle assay, as described previously (25). The half-maximal effective concentrations (EC50) were determined by nonlinear regression of data converted to percent cell death using SigmaPlot (Systat Software, San Jose, CA). Statistical significance of the fold changes for the mutants compared to the wild-type control was calculated with the two-tailed Student's t test (P < 0.05).
Determination of viral replication fitness by growth competition assay.
Viral growth competitions were performed by coculturing competing recombinant xxLAI viruses in MT-2 cells for 9 days in the presence or absence of ARV inhibitors, as described previously (18, 20, 21). MultiCode RTx PCR (Luminex, Austin, TX) was performed on extracted viral RNA from days 0, 3, 6, and 9 as described previously with allele-specific forward and reverse primers (20, 21, 26). Percentages of competing viruses were determined from standard curves generated by SigmaPlot. Quantitative estimates of relative fitness (RF) were calculated using the “1 + s” equation (18, 27). An RF value of 1.00 indicates that the two viruses grew with equivalent fitnesses, a value of <1.00 indicates less efficient growth of the second virus relative to that of the first virus competitor, and a value of >1.00 indicates enhanced fitness of the second virus relative to that of the first virus competitor. The mean RF values from at least three independent competition experiments are reported, and statistical significances of RF values from the replicates of a competition experiment compared to the wild-type versus wild-type control were calculated by the two-tailed Student's t test (P ≤ 0.01).
RESULTS
NRTI and INSTI resistance substitutions only reduce phenotypic susceptibility to drugs in-class.
Viruses containing the RT-K65R, RT-M184V, IN-Q148R, and IN-N155H substitutions, alone and as RT-IN combinations, were evaluated for phenotypic susceptibility to INSTIs, selected NRTIs, and a representative inhibitor from the NNRTI and PI drug classes. All viruses containing the IN-Q148R or IN-N155H substitutions demonstrated reduced susceptibility to EVG and RAL (Table 1). Resistance to EVG ranged from 101- to 137-fold for IN-Q148R-containing viruses and 40- to 55-fold for IN-N155H-containing viruses. Resistance to RAL ranged from 44- to 46-fold for the IN-Q148R mutants and 19- to 23-fold for the IN-N155H mutants. Thus, the reductions in INSTI susceptibility were consistent for each IN substitution, regardless of RT sequence. Viruses with IN-Q148R or IN-N155H substitutions maintained sensitivity to the newest INSTI DTG (fold change compared to wild type < 2.5). The RT-K65R and RT-M184V mutants remained fully susceptible to all INSTIs tested.
TABLE 1.
Phenotypic susceptibilities of HIV-1xxLAI with NRTI and INSTI drug resistance substitutions to antiretroviral inhibitors
| HIV-1xxLAI genotype | Fold change in EC50 relative to that of the wild typea |
|||||||
|---|---|---|---|---|---|---|---|---|
| INSTIs |
NRTIs |
PI |
NNRTI |
|||||
| EVG | RAL | DTG | TFV | FTC | AZT | DRV | EFV | |
| RT-K65R | 1.1 ± 0.3 | 1.0 ± 0.2 | 1.1 ± 0.1 | 3.6 ± 0.5 | 23 ± 9.3 | 0.6 ± 0.1 | 1.1 ± 0.2 | 0.8 ± 0.1 |
| RT-M184V | 0.8 ± 0.3 | 0.9 ± 0.1 | 0.9 ± 0.2 | 0.6 ± 0.1 | >1,000 | 0.4 ± 0.2 | 0.9 ± 0.1 | 0.7 ± 0.4 |
| IN-Q148R | 137 ± 84 | 46 ± 9.9 | 1.8 ± 0.4 | 1.0 ± 0.3 | 1.2 ± 0.3 | 0.7 ± 0.5 | 0.9 ± 0.1 | 1.0 ± 0.6 |
| RT-M184V + IN-Q148R | 132 ± 49 | 46 ± 15 | 1.7 ± 0.2 | 0.8 ± 0.4 | >1,000 | 0.5 ± 0.2 | 1.0 ± 0.3 | 0.8 ± 0.6 |
| RT-K65R/M184V + IN-Q148R | 101 ± 29 | 44 ± 14 | 1.4 ± 0.4 | 3.4 ± 0.6 | >1,000 | 0.6 ± 0.3 | 0.9 ± 0.1 | 0.6 ± 0.1 |
| IN-N155H | 40 ± 34 | 19 ± 3.0 | 1.9 ± 0.6 | 0.9 ± 0.3 | 1.4 ± 0.6 | 0.9 ± 0.4 | 0.9 ± 0.1 | 0.9 ± 0.3 |
| RT-M184V + IN-N155H | 55 ± 25 | 22 ± 5.8 | 2.1 ± 1.0 | 0.8 ± 0.6 | >1,000 | 0.4 ± 0.3 | 0.9 ± 0.1 | 0.7 ± 0.4 |
| RT-K65R/M184V + IN-N155H | 48 ± 9.5 | 23 ± 5.5 | 1.7 ± 0.3 | 3.3 ± 1.0 | >1,000 | 0.6 ± 0.4 | 0.9 ± 0.1 | 0.6 ± 0.1 |
Data represent the geometric means from at least 3 independent experiments. The EC50s for the wild type were 2.3 nM for EVG, 6.4 nM for RAL, 1.0 nM for DTG, 4.1 μM for TFV, 0.7 μM for FTC, 0.2 μM for AZT, 6.3 nM for DRV, and 1.7 nM for EFV. Fold changes were calculated by setting the mean wild-type EC50 at 1, and all fold change values of >2.5 (in bold) demonstrated statistically significant differences using a two-tailed Student's t test (P < 0.05).
Next, phenotypic susceptibilities to NRTIs (FTC, TFV, and AZT) were assessed (Table 1). The presence of the RT-M184V substitution resulted in high levels of resistance to FTC (>1,000-fold). The RT-K65R mutant had reduced susceptibilities to TFV (3.6-fold) and FTC (23-fold). The two triple mutants with RT-K65R/M184V + IN-Q148R and RT-K65R/M184V + IN-N155H substitutions also had moderately reduced susceptibilities to TFV (3.4- and 3.3-fold, respectively), while the rest of the viruses without RT-K65R remained fully susceptible to TFV. All viruses with the RT-K65R and/or RT-M184V substitutions had slightly increased sensitivities to AZT, consistent with our previous study (≤0.6-fold) (18). The IN-Q148R and IN-N155H single mutants were fully susceptible to all NRTIs tested. Furthermore, all mutant viruses maintained full sensitivities to the NNRTI EFV and the PI DRV. Overall, viral susceptibility to INSTIs and NRTIs was affected only by substitutions in the corresponding target enzymes, with no cross-class resistance observed.
NRTI and INSTI resistance substitutions diminish viral fitness in the absence of drug pressure.
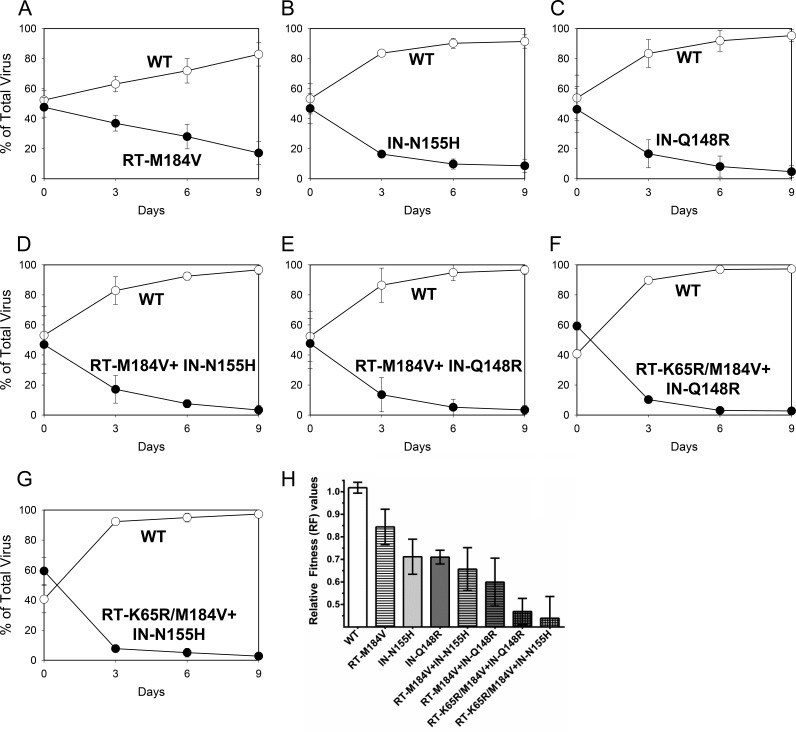
Pairwise growth competitions were performed to evaluate the relative fitnesses of viruses containing RT-K65R, RT-M184V, IN-Q148R, and IN-N155H. The replication efficiency of each virus was determined by calculating relative fitness (RF) values (Table 2). Mutant viral fitness was first evaluated by direct competition with the wild type (Fig. 1). The RT-M184V, IN-N155H, and IN-Q148R mutants all demonstrated reduced fitnesses relative to that of the wild type. The combination of RT-M184V with either IN-N155H or IN-Q148R resulted in a greater fitness defect than any single substitution alone. Previously we reported that the addition of the RT-K65R substitution to wild-type and mutant viruses contributed to cumulative reductions in viral fitness in the absence of drugs (18). In this study, we similarly observed that mutants containing RT-K65R/M184V combined with IN-N155H or IN-Q148R showed the greatest reductions in viral fitness relative to that of the wild type. Overall, viral fitness of RT and/or IN mutants in competition with the wild type diminished in the following genotypic order: wild type > RT-M184V ≥ IN-N155H ≈ IN-Q148R ≥ RT-M184V + IN-N155H ≥ RT-M184V + IN-Q148R ≥ RT-K65R/M184V + IN-Q148R ≈ RT-K65R/M184V + IN-N155H.
TABLE 2.
Relative fitnesses of wild-type and site-directed mutant HIV-1xxLAI in growth competition assays
| Competition condition and genotype of competing viruses | Relative fitness valuea | P valueb | Fitness interpretation |
|---|---|---|---|
| Without drug | |||
| WT vs WTc | 1.02 ± 0.02 | WT = WT | |
| WT vs RT-M184V | 0.84 ± 0.08 | 0.008 | WT > RT-M184V |
| WT vs IN-N155H | 0.71 ± 0.08 | 0.001 | WT > IN-N155H |
| WT vs IN-Q148R | 0.71 ± 0.03 | <0.001 | WT > IN-Q148R |
| WT vs RT-M184V + IN-N155H | 0.66 ± 0.1 | <0.001 | WT > RT-M184V + IN-N155H |
| WT vs RT-M184V + IN-Q148R | 0.60 ± 0.1 | <0.001 | WT > RT-M184V + IN-Q148R |
| WT vs RT-K65R/M184V + IN-Q148R | < 0.50 ± 0.1 | <0.001 | WT > RT-K65R/M184V + IN-Q148R |
| WT vs RT-K65R/M184V + IN-N155H | < 0.50 ± 0.2 | <0.001 | WT > RT-K65R/M184V + IN-N155H |
| IN-N155H vs IN-Q148R | 0.97 ± 0.06 | 0.164 | IN-N155H ≈ IN-Q148R |
| RT-M184V vs RT-M184V + IN-Q148R | 0.75 ± 0.05 | <0.001 | RT-M184V > RT-M184V + IN-Q148R |
| RT-M184V vs RT-M184V + IN-N155H | 0.76 ± 0.01 | <0.001 | RT-M184V > RT-M184V + IN-N155H |
| IN-Q148R vs RT-M184V + IN-Q148R | 0.93 ± 0.09 | 0.111 | IN-Q148R ≥ RT-M184V + IN-Q148R |
| IN-N155H vs RT-M184V + IN-N155H | 0.94 ± 0.07 | 0.091 | IN-N155H ≥ RT-M184V + IN-N155H |
| With drug | |||
| WT vs IN-Q148R (0.5 nM EVG) | 0.76 ± 0.02 | <0.001 | WT > IN-Q148R |
| WT vs IN-Q148R (2 nM EVG) | 0.95 ± 0.04 | 0.038 | WT ≈ IN-Q148R |
| WT vs IN-Q148R (10 nM EVG) | 2.09 ± 0.1 | <0.001 | WT < IN-Q148R |
| WT vs IN-N155H (0.5 nM EVG) | 0.85 ± 0.04 | 0.001 | WT > IN-N155H |
| WT vs IN-N155H (2 nM EVG) | 1.10 ± 0.05 | 0.043 | WT ≤ IN-N155H |
| WT vs IN-N155H (10 nM EVG) | 2.03 ± 0.5 | 0.010 | WT < IN-N155H |
| IN-N155H vs IN-Q148R (1 nM EVG) | 1.07 ± 0.06 | 0.178 | IN-N155H ≤ IN-Q148R |
| IN-N155H vs IN-Q148R (10 nM EVG) | 0.83 ± 0.05 | 0.001 | IN-N155H > IN-Q148R |
| IN-N155H vs IN-Q148R (50 nM EVG) | 1.32 ± 0.4 | 0.194 | IN-N155H ≤ IN-Q148R |
| IN-N155H vs IN-Q148R (100 nM EVG) | 1.77 ± 0.1 | <0.001 | IN-N155H < IN-Q148R |
| RT-M184V vs RT-M184V + IN-Q148R (0.5 nM EVG, 1 nM FTC) | 0.82 ± 0.05 | 0.001 | RT-M184V > RT-M184V + IN-Q148R |
| RT-M184V vs RT-M184V + IN-Q148R (10 nM EVG, 100 nM FTC) | 1.38 ± 0.09 | 0.001 | RT-M184V < RT-M184V + IN-Q148R |
| RT-M184V vs RT-M184V + IN-N155H (0.5 nM EVG, 1 nM FTC) | 0.84 ± 0.1 | 0.024 | RT-M184V ≥ RT-M184V + IN-N155H |
| RT-M184V vs RT-M184V + IN-N155H (10 nM EVG, 100 nM FTC) | 1.57 ± 0.2 | 0.006 | RT-M184V < RT-M184V + IN-N155H |
| IN-Q148R vs RT-M184V + IN-Q148R (1 nM EVG, 1 nM FTC) | 0.83 ± 0.1 | 0.037 | IN-Q148R ≥ RT-M184V + IN-Q148R |
| IN-Q148R vs RT-M184V + IN-Q148R (100 nM EVG, 100 nM FTC) | 1.61 ± 0.3 | 0.010 | IN-Q148R < RT-M184V + IN-Q148R |
| IN-N155H vs RT-M184V + IN-N155H (1 nM EVG, 1 nM FTC) | 0.86 ± 0.1 | 0.029 | IN-N155H ≥ RT-M184V + IN-N155H |
| IN-N155H vs RT-M184V + IN-N155H (100 nM EVG, 100 nM FTC) | 1.38 ± 0.1 | 0.003 | IN-N155H < RT-M184V + IN-N155H |
The relative fitness (RF) value of the mutant in competition with the wild type was calculated as follows: (1 + s) = exp {(1/t) × ln[(Mt/Wt) × (Mt0/Wt0)]}, where s is the selection coefficient; t is the time (in days); Mt and Mt0 are the fractions of mutant virus initially and at the time of measurement, respectively; and Wt and Wt0 are the fractions of wild-type virus initially and at the time of measurement, respectively (27). Mutant-versus-mutant competitions were analyzed using the same equation. The data represent the means and standard deviations from at least 3 independent experiments.
P values were determined using a two-tailed Student's t test comparing the competitions to the wild-type-versus-wild-type competition.
A control experiment was performed to verify that isogenic HIV-1 recombinants differing only in their sequence tags would grow with equivalent fitnesses. WT, wild type.
FIG 1.
Growth competitions of wild-type HIV-1 (WT; open circles) versus the RT-M184V (A), IN-N155H (B), IN-Q148R (C), RT-M184V + IN-N155H (D), RT-M184V + IN-Q148R (E), RT-K65R/M184V + IN-Q148R (F), and RT-K65R/M184V + IN-N155H (G) mutants (closed circles). Data are averages from 3 independent competition experiments, with standard deviations. (H) Mean relative fitness (RF; 1 + s) values of viruses (genotype indicated) in competition with the WT. Asterisks indicate statistically significant differences for the mutant-versus-WT competitions compared to the WT-versus-WT control competitions using a two-tailed Student's t test (P < 0.01).
Direct growth competitions between mutant viruses were also performed to further characterize the fitness of viruses with RT and IN substitutions. The IN-Q148R mutant grew with a fitness almost equivalent to that of the IN-N155H mutant (Fig. 2A). The RT-M184V + IN-Q148R and RT-M184V + IN-N155H double mutants were both less fit than the RT-M184V single mutant (Fig. 3A and D). Additionally, the RT-M184V + IN-Q148R mutant was slightly less fit than the IN-Q148R mutant (Fig. 3G), and the RT-M184V + IN-N155H mutant was slightly less fit than the IN-N155H mutant (Fig. 3J). Overall, the mutant-versus-mutant competitions confirmed the results from the wild-type-versus-mutant competitions, indicating that the IN-Q148R and IN-N155H substitutions diminish viral fitness similarly and to a greater degree than the RT-M184V substitution.
FIG 2.
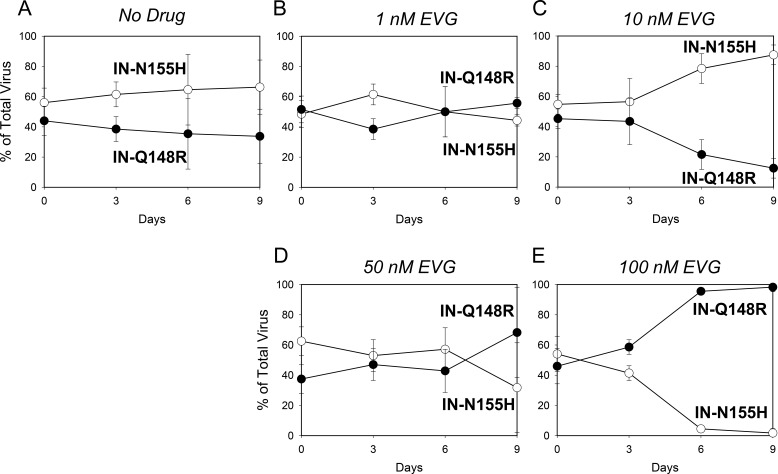
Growth competitions of the IN-N155H mutant (open circles) versus the IN-Q148R mutant (closed circles) in the presence of no drug (A), 1 nM EVG (B), 10 nM EVG (C), 50 nM EVG (D), and 100 nM EVG (E). Data are averages from 3 independent competition experiments, with standard deviations. Mean relative fitness (RF) values for the IN-Q148R mutant compared to the IN-N155H mutant are shown in Table 2.
FIG 3.
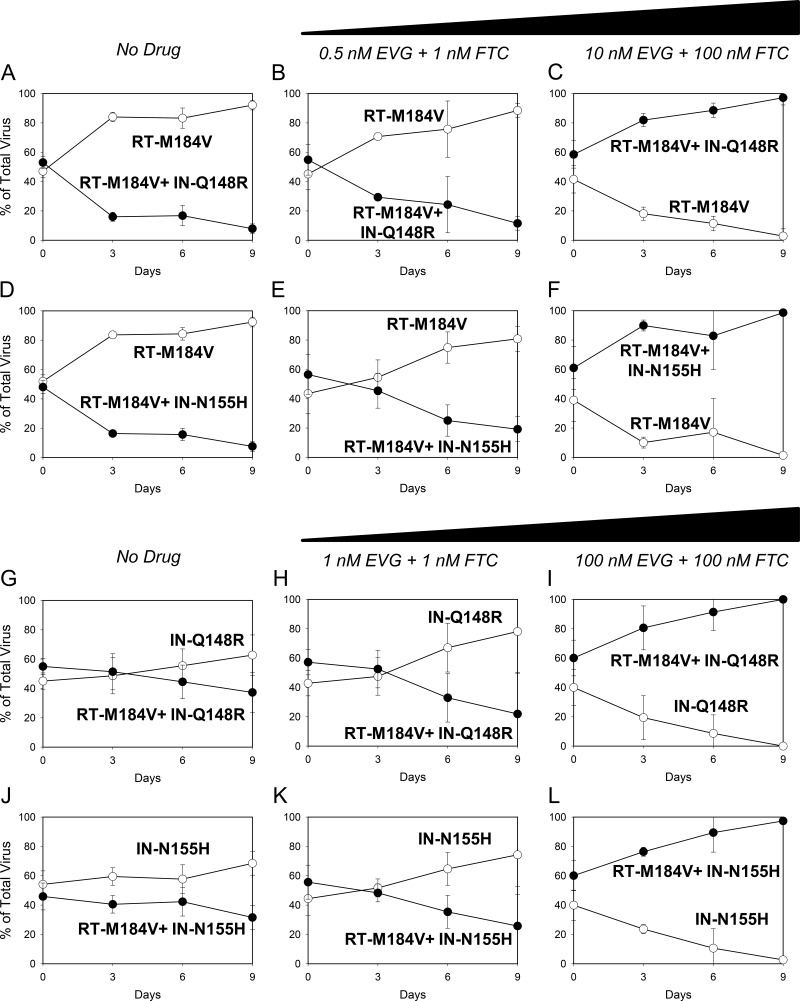
(A to F) Growth competitions of the RT-M184V mutant versus the RT-M184V + IN-Q148R and RT-M184V + IN-N155H mutants in the presence of no drug (A and D, respectively), 0.5 nM EVG + 1 nM FTC (B and E, respectively), and 10 nM EVG + 100 nM FTC (C and F, respectively). (G to L) Growth competitions of the IN-Q148R mutant versus the RT-M184V + IN-Q148R mutant and the IN-N155H versus RT-M184V + IN-N155H mutant in the presence of no drug (G and J, respectively), 1 nM EVG plus 1 nM FTC (H and K, respectively), and 100 nM EVG plus 100 nM FTC (I and L, respectively) Data are averages from 3 independent competition experiments, with standard deviations. Mean relative fitness (RF) values for the double mutants (closed circles) compared to the single mutants (open circles) are shown in Table 2.
Resistance compensates for viral fitness in the presence of antiretroviral drugs.
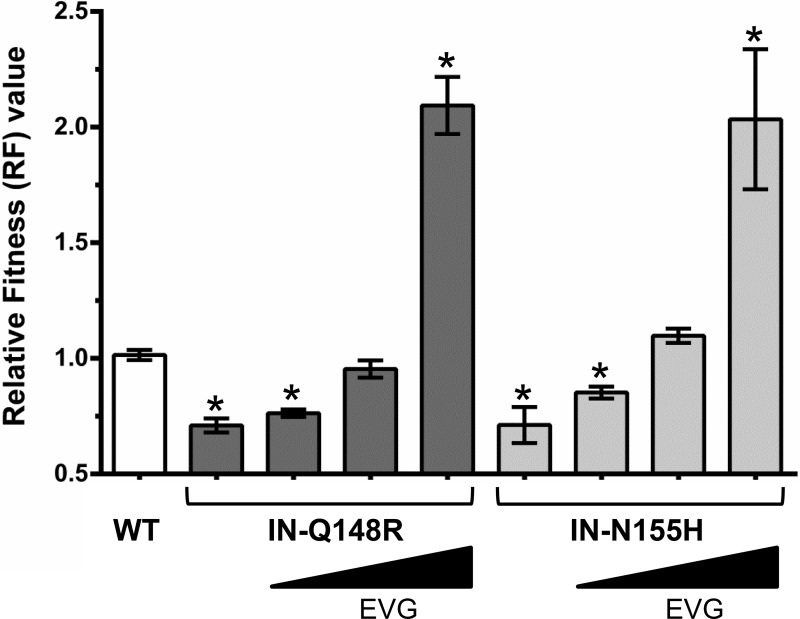
To determine how resistance influences viral fitness, growth competitions were performed in the presence of ARV drugs at concentrations approaching physiologic conditions. First, IN mutants were grown in competition against the wild type in the presence of EVG (Fig. 4). With 0.5 nM EVG present, the IN-Q148R and IN-N155H mutants remained less fit than the wild type. At 2 nM EVG, the IN-Q148R mutant and wild type grew similarly, while the IN-N155H mutant began to slightly outcompete the wild type. At 10 nM EVG, the IN-Q148R and IN-N155H mutants outgrew the wild type.
FIG 4.
Relative fitness (RF) values of wild-type and mutant HIV-1 in competition with the wild type in the absence and presence of increasing EVG concentrations (0.5 nM, 2 nM, and 10 nM). Asterisks indicate statistically significant differences for the mutant-versus-WT competitions compared to the WT-versus-WT control competitions using a two-tailed Student's t test (P < 0.01).
Next, competitions between the IN-Q148R and IN-N155H mutants were conducted in the presence of EVG (Fig. 2B to E). At 1 nM EVG, the fitness of the IN-Q148R mutant continued to be similar to that of the IN-N155H mutant. At 10 nM EVG, the IN-Q148R mutant had reduced fitness relative to the IN-N155H mutant. Interestingly, the IN-Q148R mutant began to outgrow the IN-N155H mutant in the competition at 50 nM EVG and then completely outcompeted the IN-N155H mutant at 100 nM EVG. Thus, despite having equivalent fitnesses in the absence of EVG, the IN-Q148R and IN-N155H mutants demonstrated fitness advantages in the presence of EVG that reflected their levels of resistance. In other words, the less resistant IN-N155H mutant grew more efficiently at a lower EVG concentration, while the more resistant IN-Q148R mutant grew more efficiently at the higher EVG concentrations.
Finally, single mutants were grown in competition against double mutants in the presence of EVG and FTC. With 0.5 nM EVG and 1 nM FTC present, the RT-M184V + IN-Q148R and RT-M184V + IN-N155H mutants grew less efficiently than the RT-M184V mutant in head-to-head competitions, resembling the results of the competitions performed in the absence of drug (Fig. 3A, B, D, and E). However, in the presence of higher drug concentrations (10 nM EVG and 100 nM FTC), both double mutants were able to outgrow the RT-M184V mutant in their respective competitions (Fig. 3C and F). Similarly, the RT-M184V + IN-Q148R mutant remained less fit than the IN-Q148R mutant in the presence of 1 nM EVG + 1 nM FTC (Fig. 3H) but was able to outcompete the IN-Q148R mutant with 100 nM EVG and 100 nM FTC present (Fig. 3I). The competitions between the RT-M184V + IN-N155H mutant versus IN-N155H mutant with EVG and FTC had similar results: the RT-M184V + IN-N155H mutant was less fit than the IN-N155H mutant at the low drug concentrations but was able to outgrow the IN-N155H mutant at the high drug concentrations (Fig. 3K and L). Overall, these results demonstrate how resistance can counteract fitness defects during antiretroviral drug pressure.
DISCUSSION
Suppressive ARV therapy prevents HIV-1 disease progression, but long-term treatment efficacy can be jeopardized by the development of drug resistance. In patients experiencing virologic failure on regimens containing EVG or RAL plus an NRTI backbone, NRTI and INSTI resistance substitutions have been found to codevelop (12–16, 28–30), suggesting potential cross-class mutational interactions. We constructed HIV-1 site-directed mutants based on the most common resistance profiles observed in EVG/COBI/FTC/TDF phase 3 studies to explore the combined effects of NRTI and INSTI resistance substitutions on viral fitness and drug resistance. We previously reported that the RT-K65R, RT-M184V, and IN-E92Q substitutions did not show any cross-class resistance or fitness compensation in the absence of drug pressure (18). We have now broadened our study to include other clinically relevant INSTI resistance substitutions, IN-Q148R and IN-N155H, in combination with RT-M184V and RT-K65R. Similar to our previous conclusions, we found that drug susceptibility depended mainly on the genotype of the target enzyme and that the RT-M184V, IN-Q148R, and IN-N155H substitutions each caused viral fitness defects that were additive but could be offset by resistance in the presence of drugs.
EVG and RAL, the first two INSTIs to gain regulatory approval for HIV-1 treatment, have complex and overlapping resistance profiles consisting of multiple primary and secondary mutations in IN (22, 31). Primary substitutions for both drugs, including IN-Q148R and IN-N155H, have previously been associated with diminished viral replicative fitness and are rarely observed in combination on the same viral genomes in vivo (9, 17, 32, 33). Instead, INSTI primary resistance mutants may coexist as distinct subpopulations that evolve over time with or without development of secondary INSTI resistance substitutions (9, 31, 34). Temporal shifts in resistance pathways from IN-N155H to IN-Q148H/K/R have been observed in patients failing RAL-containing regimens, likely due to the selective advantage of more resistant IN-Q148H/K/R mutants under continued drug pressure (35–38). Our data support these findings in the context of EVG, with the less resistant IN-N155H mutant appearing to have a fitness advantage over the IN-Q148R mutant at low EVG concentrations and the reverse occurring at high EVG concentrations. In the absence of drug, the IN-Q148R and IN-N155H mutants had almost equivalently reduced fitnesses relative to that of the wild type, similar to previous reports (1, 22, 38). IN-Q148R and IN-N155H resulted in greater reductions in viral fitness than did the IN-E92Q substitution previously studied, which may partially explain why the IN-E92Q substitution developed more frequently than the IN-Q148R or IN-N155H substitution in the EVG/COBI/FTC/TDF-treated virologic-failure patients (16).
Unlike the resistance patterns observed during clinical trials of EVG and RAL, clinical trials of ARV-naive patients treated with DTG have not yielded development of INSTI and NRTI resistance substitutions in combination (39–43). The significant fitness defect caused by the primary DTG resistance substitution, IN-R263K, and lack of compensatory mutations to restore viral fitness have been proposed to play a role in preventing resistance development to DTG-containing regimens (44–49). Furthermore, the RT-M184V and RT-M184I substitutions have been shown to further reduce viral fitness, but not result in cross-class drug resistance, when combined with IN-R263K (50), similar to our findings presented here. However, the RT-M184I substitution was able to develop in addition to IN-R263K under the dose-escalating selective pressure of lamivudine (51), suggesting that at least in vitro, HIV-1 can accommodate the fitness defects of IN-R263K together with NRTI resistance substitutions. Thus, factors other than viral fitness may be involved in the very low rates of resistance development in clinical trials of DTG-based regimens. Currently, DTG and the NNRTI rilpivirine are being investigated as a 2-drug regimen for maintenance of virologic suppression. Given the recent findings that certain NNRTI and INSTI resistance substitutions may have cross-class interactions leading to enhanced drug resistance (3), it will be interesting to see if resistance development while on this DTG-based regimen remains low.
Overall, NRTI and INSTI resistance substitutions are associated with reduced viral fitness compared to that of wild-type HIV in the absence of drug. The substitutions RT-K65R, RT-M184V, IN-Q148R, and IN-N155H did not exhibit cross-class resistance. Double mutants containing RT-M184V with IN-Q148R or IN-N155H demonstrated a competitive advantage in the presence of FTC and EVG, reflecting the contribution of resistance to viral fitness in the setting of multidrug pressure.
REFERENCES
- 1.Hu Z, Kuritzkes DR. 2010. Effect of raltegravir resistance mutations in HIV-1 integrase on viral fitness. J Acquir Immune Defic Syndr 55:148–155. doi: 10.1097/QAI.0b013e3181e9a87a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delelis O, Malet I, Na L, Tchertanov L, Calvez V, Marcelin AG, Subra F, Deprez E, Mouscadet JF. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res 37:1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Z, Kuritzkes DR. 2014. Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors. J Virol 88:9268–9276. doi: 10.1128/JVI.00695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buzón MJ, Dalmau J, Puertas MC, Puig J, Clotet B, Martinez-Picado J. 2010. The HIV-1 integrase genotype strongly predicts raltegravir susceptibility but not viral fitness of primary virus isolates. AIDS 24:17–25. doi: 10.1097/QAD.0b013e328331c81e. [DOI] [PubMed] [Google Scholar]
- 5.Weber J, Rose JD, Vazquez AC, Winner D, Margot N, McColl DJ, Miller MD, Quinones-Mateu ME. 2013. Resistance mutations outside the integrase coding region have an effect on human immunodeficiency virus replicative fitness but do not affect its susceptibility to integrase strand transfer inhibitors. PLoS One 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J Virol 81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hehl EA, Joshi P, Kalpana GV, Prasad VR. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J Virol 78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol 73:2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winters MA, Lloyd RM Jr, Shafer RW, Kozal MJ, Miller MD, Holodniy M. 2012. Development of elvitegravir resistance and linkage of integrase inhibitor mutations with protease and reverse transcriptase resistance mutations. PLoS One 7:e40514. doi: 10.1371/journal.pone.0040514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina JM, Lamarca A, Andrade-Villanueva J, Clotet B, Clumeck N, Liu YP, Zhong L, Margot N, Cheng AK, Chuck SL. 2012. Efficacy and safety of once daily elvitegravir versus twice daily raltegravir in treatment-experienced patients with HIV-1 receiving a ritonavir-boosted protease inhibitor: randomised, double-blind, phase 3, non-inferiority study. Lancet Infect Dis 12:27–35. doi: 10.1016/S1473-3099(11)70249-3. [DOI] [PubMed] [Google Scholar]
- 11.Charpentier C, Karmochkine M, Laureillard D, Tisserand P, Belec L, Weiss L, Si-Mohamed A, Piketty C. 2008. Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med 9:765–770. doi: 10.1111/j.1468-1293.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- 12.White KL, Kulkarni R, McColl DJ, Rhee MS, Szwarcberg J, Cheng AK, Miller MD. 16 October 2014. Week 144 resistance analysis of elvitegravir/cobicistat/emtricitabine/tenofovir DF versus efavirenz/emtricitabine/tenofovir DF in antiretroviral-naive patients. Antivir Ther doi: 10.3851/IMP2885. [DOI] [PubMed] [Google Scholar]
- 13.Kulkarni R, Abram ME, McColl DJ, Barnes T, Fordyce MW, Szwarcberg J, Cheng AK, Miller MD, White KL. 2014. Week 144 resistance analysis of elvitegravir/cobicistat/emtricitabine/tenofovir df versus atazanavir+ritonavir+emtricitabine/tenofovir DF in antiretroviral-naive patients. HIV Clin Trials 15:218–230. doi: 10.1310/hct1505-218. [DOI] [PubMed] [Google Scholar]
- 14.Clumeck N, Molina JM, Henry K, Gathe J, Rockstroh JK, Dejesus E, Wei X, White K, Fordyce MW, Rhee MS, Szwarcberg J. 2014. A randomized, double-blind comparison of single-tablet regimen elvitegravir/cobicistat/emtricitabine/tenofovir DF vs ritonavir-boosted atazanavir plus emtricitabine/tenofovir DF for initial treatment of HIV-1 infection: analysis of week 144 results. J Acquir Immune Defic Syndr 65:e121–e124. doi: 10.1097/QAI.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 15.Wohl DA, Cohen C, Gallant JE, Mills A, Sax PE, Dejesus E, Zolopa A, Liu HC, Plummer A, White KL, Cheng AK, Rhee MS, Szwarcberg J. 2014. A randomized, double-blind comparison of single-tablet regimen elvitegravir/cobicistat/emtricitabine/tenofovir DF versus single-tablet regimen efavirenz/emtricitabine/tenofovir DF for initial treatment of HIV-1 infection: analysis of week 144 results. J Acquir Immune Defic Syndr 65:e118–e121. doi: 10.1097/QAI.0000000000000057. [DOI] [PubMed] [Google Scholar]
- 16.White K, Kulkarni R, Miller MD. 2015. Analysis of early resistance development at the first failure timepoint in elvitegravir/cobicistat/emtricitabine/tenofovir disoproxil fumarate-treated patients. J Antimicrob Chemother 70:2632–2638. doi: 10.1093/jac/dkv149. [DOI] [PubMed] [Google Scholar]
- 17.White K, Kulkarni R, Abram M, Rhee M, Fordyce M, Szwarcberg J, Miller M. 2014. Longitudinal resistance analysis of the phase 3 EVG/COBI/FTC/TDF studies, poster O_12, p 1 12th Eur Meet HIV Hepatitis Treatment Strategies Antiviral Drug Resistance, Barcelona, Spain. [Google Scholar]
- 18.Andreatta KN, Goodman DD, Miller MD, White KL. 2015. Reduced viral fitness and lack of cross-class resistance with integrase strand transfer inhibitor and nucleoside reverse transcriptase inhibitor resistance mutations. Antimicrob Agents Chemother 59:3441–3449. doi: 10.1128/AAC.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi C, Mellors JW. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother 41:2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svarovskaia ES, Feng JY, Margot NA, Myrick F, Goodman D, Ly JK, White KL, Kutty N, Wang R, Borroto-Esoda K, Miller MD. 2008. The A62V and S68G mutations in HIV-1 reverse transcriptase partially restore the replication defect associated with the K65R mutation. J Acquir Immune Defic Syndr 48:428–436. doi: 10.1097/QAI.0b013e31817bbe93. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, Svarovskaia E, Miller MD, White KL. 2012. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr 59:47–54. doi: 10.1097/QAI.0b013e31823aca74. [DOI] [PubMed] [Google Scholar]
- 22.Abram ME, Hluhanich RM, Goodman DD, Andreatta KN, Margot NA, Ye L, Niedziela-Majka A, Barnes TL, Novikov N, Chen X, Svarovskaia ES, McColl DJ, White KL, Miller MD. 2013. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob Agents Chemother 57:2654–2663. doi: 10.1128/AAC.02568-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavanaugh J, Svarovskaia ES, Goodman DD, Myrick F, Hluhanich R, McColl DJ, Miller MD, Borroto-Esoda K. 2007. Reduction in HIV-1 fitness associated with the E92Q integrase inhibitor resistance mutation: accumulation of additional IN mutations increases resistance to elvitegravir without rescuing viral fitness, poster 32 8th Annu Symp Antiviral Drug Resistance Targets Mechanisms, Richmond, VA. [Google Scholar]
- 24.Hierholzer JC, Killington RA. 1996. Virus isolation and quantitation, p 25–46. In Mahy BWJ, Kangro HO (ed), Virology methods manual. Academic Press Ltd, Waltham, Massachusetts. [Google Scholar]
- 25.Margot NA, Hluhanich RM, Jones GS, Andreatta KN, Tsiang M, McColl DJ, White KL, Miller MD. 2012. In vitro resistance selections using elvitegravir, raltegravir, and two metabolites of elvitegravir M1 and M4. Antiviral Res 93:288–296. doi: 10.1016/j.antiviral.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Svarovskaia ES, Moser MJ, Bae AS, Prudent JR, Miller MD, Borroto-Esoda K. 2006. MultiCode-RTx real-time PCR system for detection of subpopulations of K65R human immunodeficiency virus type 1 reverse transcriptase mutant viruses in clinical samples. J Clin Microbiol 44:4237–4241. doi: 10.1128/JCM.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Huang Y, Dykes C, Liu D, Ma J, Perelson AS, Demeter LM. 2006. Modeling and estimation of replication fitness of human immunodeficiency virus type 1 in vitro experiments by using a growth competition assay. J Virol 80:2380–2389. doi: 10.1128/JVI.80.5.2380-2389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeJesus E, Rockstroh JK, Lennox JL, Saag MS, Lazzarin A, Zhao J, Wan H, Rodgers AJ, Walker ML, Miller M, DiNubile MJ, Nguyen BY, Teppler H, Leavitt R, Sklar P. 2012. Efficacy of raltegravir versus efavirenz when combined with tenofovir/emtricitabine in treatment-naive HIV-1-infected patients: week-192 overall and subgroup analyses from STARTMRK. HIV Clin Trials 13:228–232. doi: 10.1310/hct1304-228. [DOI] [PubMed] [Google Scholar]
- 29.Rockstroh JK, DeJesus E, Lennox JL, Yazdanpanah Y, Saag MS, Wan H, Rodgers AJ, Walker ML, Miller M, DiNubile MJ, Nguyen BY, Teppler H, Leavitt R, Sklar P. 2013. Durable efficacy and safety of raltegravir versus efavirenz when combined with tenofovir/emtricitabine in treatment-naive HIV-1-infected patients: final 5-year results from STARTMRK. J Acquir Immune Defic Syndr 63:77–85. doi: 10.1097/QAI.0b013e31828ace69. [DOI] [PubMed] [Google Scholar]
- 30.Eron JJ Jr, Rockstroh JK, Reynes J, Andrade-Villanueva J, Ramalho-Madruga JV, Bekker LG, Young B, Katlama C, Gatell-Artigas JM, Arribas JR, Nelson M, Campbell H, Zhao J, Rodgers AJ, Rizk ML, Wenning L, Miller MD, Hazuda D, DiNubile MJ, Leavitt R, Isaacs R, Robertson MN, Sklar P, Nguyen BY. 2011. Raltegravir once daily or twice daily in previously untreated patients with HIV-1: a randomised, active-controlled, phase 3 non-inferiority trial. Lancet Infect Dis 11:907–915. doi: 10.1016/S1473-3099(11)70196-7. [DOI] [PubMed] [Google Scholar]
- 31.Mbisa JL, Martin SA, Cane PA. 2011. Patterns of resistance development with integrase inhibitors in HIV. Infect Drug Resist 4:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol 83:11440–11446. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malet I, Delelis O, Soulie C, Wirden M, Tchertanov L, Mottaz P, Peytavin G, Katlama C, Mouscadet JF, Calvez V, Marcelin AG. 2009. Quasispecies variant dynamics during emergence of resistance to raltegravir in HIV-1-infected patients. J Antimicrob Chemother 63:795–804. doi: 10.1093/jac/dkp014. [DOI] [PubMed] [Google Scholar]
- 34.Canducci F, Sampaolo M, Marinozzi MC, Boeri E, Spagnuolo V, Galli A, Castagna A, Lazzarin A, Clementi M, Gianotti N. 2009. Dynamic patterns of human immunodeficiency virus type 1 integrase gene evolution in patients failing raltegravir-based salvage therapies. AIDS 23:455–460. doi: 10.1097/QAD.0b013e328323da60. [DOI] [PubMed] [Google Scholar]
- 35.Quercia R, Dam E, Perez-Bercoff D, Clavel F. 2009. Selective-advantage profile of human immunodeficiency virus type 1 integrase mutants explains in vivo evolution of raltegravir resistance genotypes. J Virol 83:10245–10249. doi: 10.1128/JVI.00894-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee R, Jensen ST, Male F, Bittinger K, Hodinka RL, Miller MD, Bushman FD. 2011. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS 25:1951–1959. doi: 10.1097/QAD.0b013e32834b34de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen HL, Charpentier C, Nguyen N, de Truchis P, Molina JM, Ruxrungtham K, Delaugerre C. 2013. Longitudinal analysis of integrase N155H variants in heavily treated patients failing raltegravir-based regimens. HIV Med 14:85–91. doi: 10.1111/j.1468-1293.2012.01039.x. [DOI] [PubMed] [Google Scholar]
- 38.Fransen S, Gupta S, Frantzell A, Petropoulos CJ, Huang W. 2012. Substitutions at amino acid positions 143, 148, and 155 of HIV-1 integrase define distinct genetic barriers to raltegravir resistance in vivo. J Virol 86:7249–7255. doi: 10.1128/JVI.06618-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raffi F, Jaeger H, Quiros-Roldan E, Albrecht H, Belonosova E, Gatell JM, Baril JG, Domingo P, Brennan C, Almond S, Min S. 2013. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis 13:927–935. doi: 10.1016/S1473-3099(13)70257-3. [DOI] [PubMed] [Google Scholar]
- 40.Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF, Richmond G, Buendia CB, Fourie J, Ramgopal M, Hagins D, Felizarta F, Madruga J, Reuter T, Newman T, Small CB, Lombaard J, Grinsztejn B, Dorey D, Underwood M, Griffith S, Min S. 2013. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382:700–708. doi: 10.1016/S0140-6736(13)61221-0. [DOI] [PubMed] [Google Scholar]
- 41.Walmsley SL, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutierrez F, Hocqueloux L, Maggiolo F, Sandkovsky U, Granier C, Pappa K, Wynne B, Min S, Nichols G. 2013. Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N Engl J Med 369:1807–1818. doi: 10.1056/NEJMoa1215541. [DOI] [PubMed] [Google Scholar]
- 42.Clotet B, Feinberg J, van Lunzen J, Khuong-Josses MA, Antinori A, Dumitru I, Pokrovskiy V, Fehr J, Ortiz R, Saag M, Harris J, Brennan C, Fujiwara T, Min S. 2014. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 383:2222–2231. doi: 10.1016/S0140-6736(14)60084-2. [DOI] [PubMed] [Google Scholar]
- 43.Raffi F, Rachlis A, Stellbrink HJ, Hardy WD, Torti C, Orkin C, Bloch M, Podzamczer D, Pokrovsky V, Pulido F, Almond S, Margolis D, Brennan C, Min S. 2013. Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet 381:735–743. doi: 10.1016/S0140-6736(12)61853-4. [DOI] [PubMed] [Google Scholar]
- 44.Quashie PK, Mesplede T, Han YS, Oliveira M, Singhroy DN, Fujiwara T, Underwood MR, Wainberg MA. 2012. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J Virol 86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wainberg MA, Mesplede T, Raffi F. 2013. What if HIV were unable to develop resistance against a new therapeutic agent? BMC Med 11:249. doi: 10.1186/1741-7015-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesplède T, Wainberg MA. 2014. Is resistance to dolutegravir possible when this drug is used in first-line therapy? Viruses 6:3377–3385. doi: 10.3390/v6093377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wares M, Mesplede T, Quashie PK, Osman N, Han Y, Wainberg MA. 2014. The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology 11:7. doi: 10.1186/1742-4690-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mesplède T, Osman N, Wares M, Quashie PK, Hassounah S, Anstett K, Han Y, Singhroy DN, Wainberg MA. 2014. Addition of E138K to R263K in HIV integrase increases resistance to dolutegravir, but fails to restore activity of the HIV integrase enzyme and viral replication capacity. J Antimicrob Chemother 69:2733–2740. doi: 10.1093/jac/dku199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mesplède T, Quashie PK, Osman N, Han Y, Singhroy DN, Lie Y, Petropoulos CJ, Huang W, Wainberg MA. 2013. Viral fitness cost prevents HIV-1 from evading dolutegravir drug pressure. Retrovirology 10:22. doi: 10.1186/1742-4690-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singhroy DN, Wainberg MA, Mesplede T. 2015. Combination of the R263K and M184I/V resistance substitutions against dolutegravir and lamivudine decreases HIV replicative capacity. Antimicrob Agents Chemother 59:2882–2885. doi: 10.1128/AAC.05181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira M, Mesplede T, Quashie PK, Moisi D, Wainberg MA. 2014. Resistance mutations against dolutegravir in HIV integrase impair the emergence of resistance against reverse transcriptase inhibitors. AIDS 28:813–819. doi: 10.1097/QAD.0000000000000199. [DOI] [PubMed] [Google Scholar]