Fig. 3.
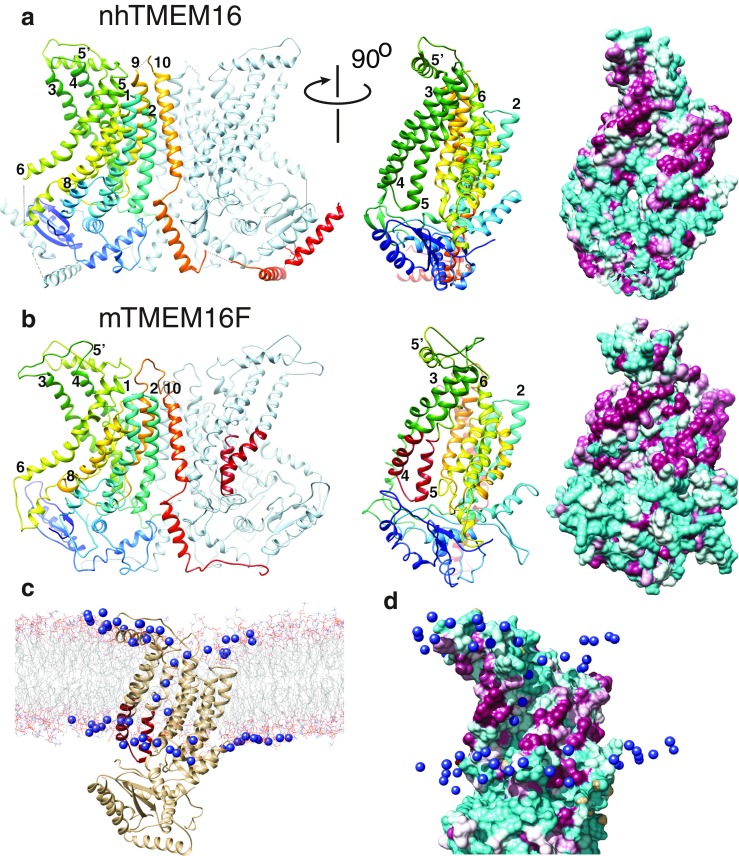
Phospholipid scrambling by TMEM16 proteins. a Crystal structure of nhTMEM16 [17] (4WIS) and b a homology model of TMEM16F based on the nhTMEM16 structure using Phyre2 [54]. One monomer is colored rainbow (blue is N-terminus, red is C-terminus) and the other is grey- light blue. Helices are numbered. Left panels: dimer viewed from the plane of the membrane. Middle panels: one monomer rotated 90° around the y-axis. The scrambling (SCRD) domain in TMEM16F in B is colored firebrick red. Right panels: molecular surfaces of the same view as the middle panel. Cyan = hydrophilic (−4.5, Kyte-Doolittle scale). Magenta = hydrophobic (4.5). c Molecular dynamics simulation of interaction of lipids with nhTMEM16 (http://sbcb.bioch.ox.ac.uk/memprotmd/beta/protein/pdbid/4WIS). A bilayer-embedded model was produced from the nhTMEM16 crystal structure through the MemProtMD protocol [104]. Lipids are shown in wire representation and the nitrogens of the choline headgroups of PtdCho molecules near the protein are shown as blue spheres. PtdCho molecules can be seen in the hydrophilic furrow and clustering near the SCRD. d Molecular surface of a close-up view of the hydrophilic furrow. The orientation is the same as c. Only the N of the PtdCho choline head groups is shown. Images were created using UCSF Chimera v. 1.10
