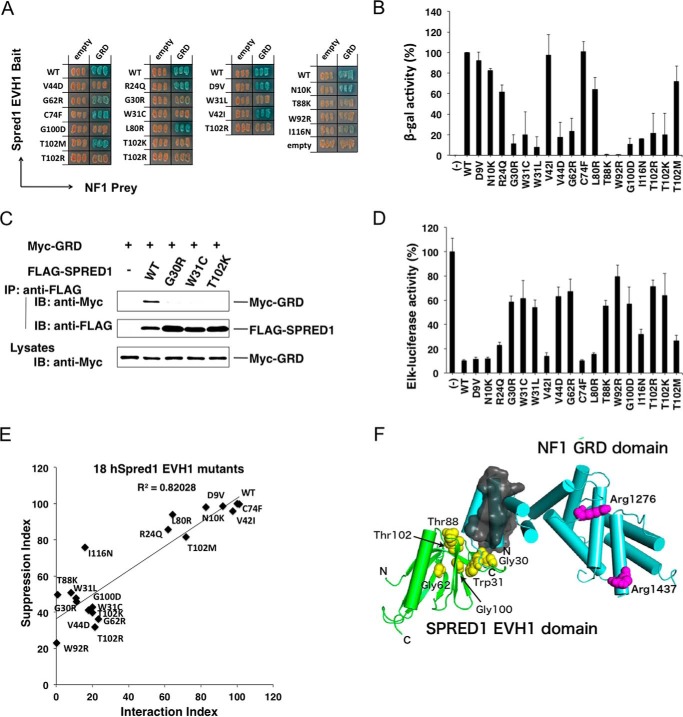
FIGURE 4.
Effects of EVH1 mutations on the EVH1-GRD interaction and ERK suppression activity. A and B, yeast strains carrying pGBKT7-hEVH1 with indicated mutations and pGADT7-GRD were restreaked on a filter paper and stained for an in situ galactosidase assay (A) and a quantitative β-gal assay (B). Error bars denote the mean ± S.D. of triplicate experiments. C, lack of interaction of the GRD with mutant EVH1 domains in 293 cells. Cells were transfected with Myc-tagged GRD and indicated FLAG-tagged SPRED1 mutants. Immunoprecipitates (IP) with anti-FLAG antibody were blotted with anti-Myc antibody. IB, immunoblot. D, ERK suppression activity by mutant SPREDs. HEK293 cells were transfected with the Elk-1 reporter system and 30 ng of mutant SPRED1 cDNAs. One day after transfection, HEK293 cells were stimulated with 50 ng/ml EGF for 6 h, and luciferase activity was then measured. E, correlation between ERK suppression activity and GRD binding activity of mutant EVH1 domains. Plotting of the EVH1-GRD interaction index and ERK suppression activity index is shown. The EVH1-GRD interaction index is calculated from B. WT EVH1 β-gal activity is standardized as 100%. ERK suppression index (calculated from D) = ((Elk-1 reporter with mutant Spred1) − (Elk-1 reporter with WT Spred1))/(Elk-1 reporter without Spred1). F, in silico modeling of the EVH1 domain (green) and GRD (light blue). The side chains of the arginine residues in charge of binding to Ras (Arg1276 and Arg1416) in GRD are shown in magenta. The N- and C-terminal helices in the GRD, which are important for EVH1 binding, are covered with a gray surface. Plausible residues in EVH1 for interaction with GRD suggested from the results of yeast two-hybrid assay and ERK suppression assay (Gly30, Trp31, Gly62, Thr88, Gly100, and Thr102) are shown in yellow.