FIGURE 10.
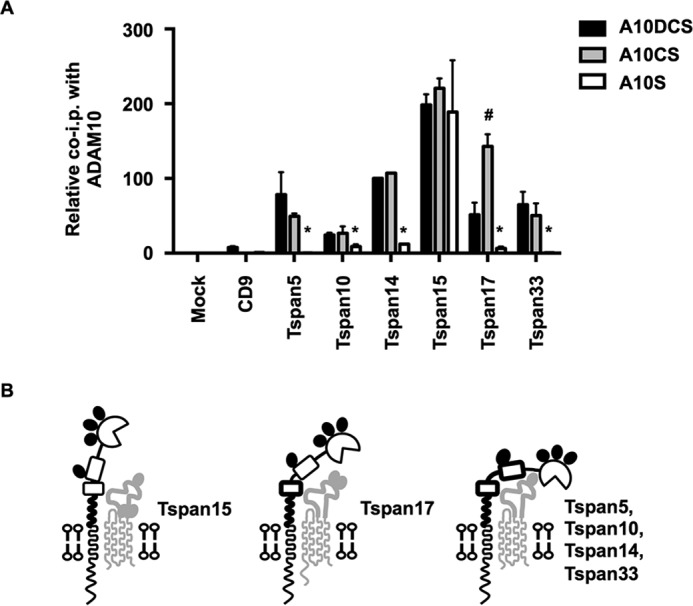
Evidence that different TspanC8s interact with ADAM10 by distinct mechanisms. A, comparison of TspanC8 co-immunoprecipitations with ADAM10 truncation constructs. Quantitation of the co-immunoprecipitations of ADAM10DCS, ADAM10CS, and ADAM10S with each tetraspanin from Fig. 9 were compared. Values were normalized using Tspan14 data from Fig. 8. All data were relative to the co-immunoprecipitation of ADAM10DCS with Tspan14, which was arbitrarily set to 100. Data were log transformed and statistical analysis was performed using a one-way ANOVA with a Dunnett's multiple comparison test comparing ADAM10CS (#, p < 0.01) or ADAM10S (*, p < 0.01) to the ADAM10DCS for each tetraspanin. Error bars represent the standard error of the mean from three experiments. B, schematic of the potential differential modes of interaction of the TspanC8s with ADAM10. Bold regions of ADAM10 represent those required for a strong interaction with the corresponding TspanC8. Note that Tspan15 has 3 N-linked glycosylation sites and Tspan17 has 2, whereas Tspan5, 10, 14, and 33 have 3, 0, 1, and 2, respectively; for the latter, Tspan14 is depicted as an example.
