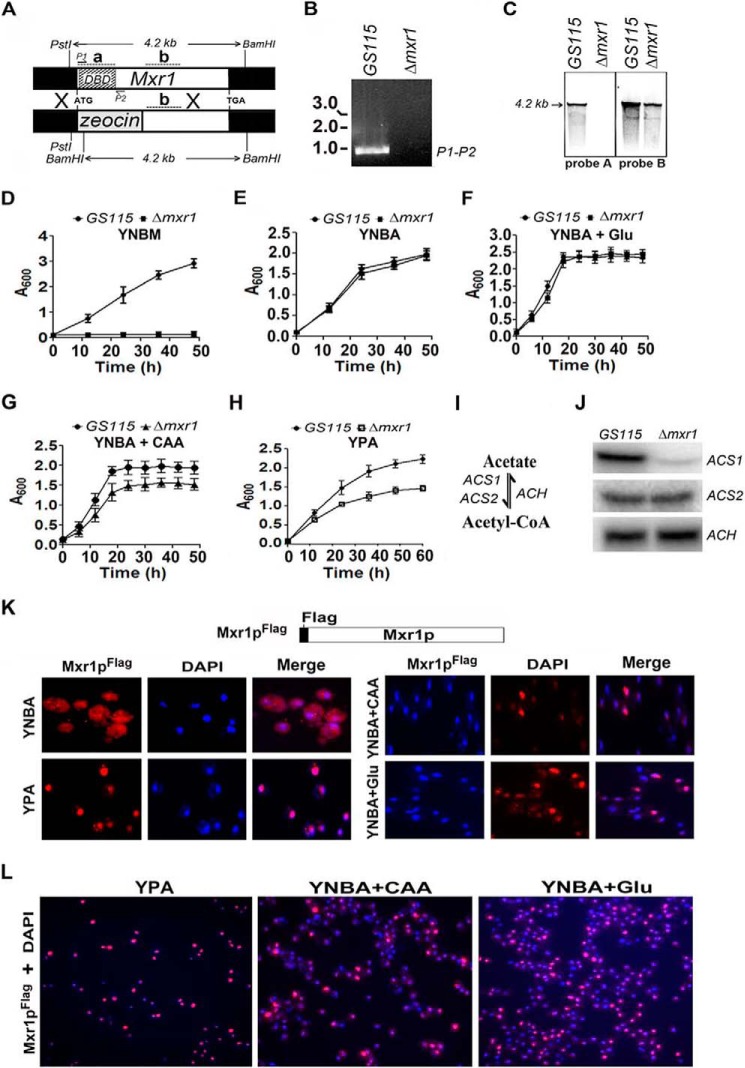
FIGURE 1.
Identification of Mxr1p as a key regulator of acetate metabolism and ACS1 expression. A, strategy for the generation of the Δmxr1 strain. The positions of the P1 and P2 primers used in PCR reaction are indicated. The restriction sites in Mxr1 and the size of the restriction fragments are indicated. Regions that hybridize to radiolabeled probes A and B in Southern blotting are indicated as a and b, respectively. B, generation of a 960-bp product by PCR amplification of genomic DNA isolated from GS115, but not Δmxr1, using the P1 and P2 primer pair, confirming the deletion of the region encoding the DNA binding domain. C, Southern blotting analysis of genomic DNA restricted with PstI and BamHI. Radiolabeled probes A and B hybridize to regions a (+1 to +554 bp) and b (+1200 to +1500 bp), located within and outside of the regions encoding the DNA binding domain, respectively. D–H, growth curves of the GS115 and Δmxr1 strains cultured in various media as indicated. Error bars indicate mean ± S.D. (n = 3). CAAs and Glu were added to YNBA medium to final concentrations of 1% and 0.5%, respectively. I, schematic of acetyl-CoA synthesis and degradation. J, Northern blotting analysis of ACS1, ACS2, and ACH. RNA was isolated from cells cultured in YPA medium for 12 h. K, immunofluorescence studies to examine the subcellular localization of Mxr1pFLAG in cells cultured in YNBA, YPA, YNBA + CAA, and YNBA + Glu media using mouse anti-FLAG antibodies and TRITC-conjugated goat anti-mouse antibodies. DAPI was used a nuclear marker. L, nuclear localization of Mxr1pFLAG in a large number of cells cultured in YPA, YNBA + CAA, and YNBA + Glu media.