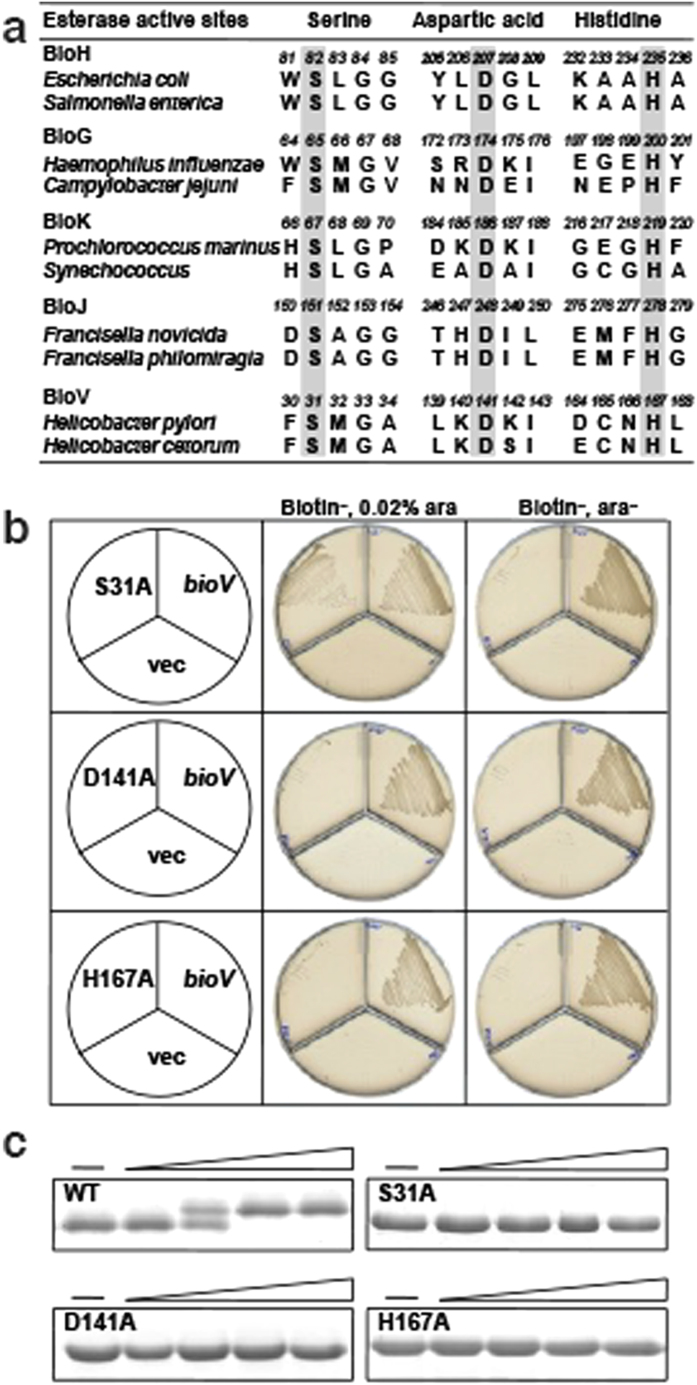
Figure 5. Identification of the BioV catalytic Ser-Asp-His triad residues.
(a) Comparison of the putative active site of BioV with the catalytic residues of putative biotin synthetic esterases. The key residues are in grey. (b) In vivo functional analyses of the triad residue BioV mutant proteins. Transformants of strain STL24 (an E. coli ∆bioH strain) were grown at 37 °C on biotin-free medium. Growth was tested in either the presence or the absence of arabinose. The strains tested were: STL24 carrying plasmids pBHK565, pBHK624, pBHK570 or pBHK571 encoding wild type bioV, or, respectively, one of the mutant derivatives, S31A, D141A or H167A. The vector plasmid (vec), pBAD24M was also included. (c) In vitro functional analyses of the triad residue BioV mutant proteins. Enzymatic activities of BioV and the single mutant S31A, D141A and H167A proteins were assayed by the conformationally sensitive electrophoretic mobility shift assay. Minus denotes no addition of BioV (or a mutant protein) whereas the triangle on the right hand represents the protein levels in a inverse dilution series (3,10, 30 and 100 nM). The enzymatic reaction (10 μl total volume) contained 100 μM pimeloyl-ACP methyl ester. The reaction mixture was separated using 18% PAGE containing 2.5 M urea.