Figure 2.
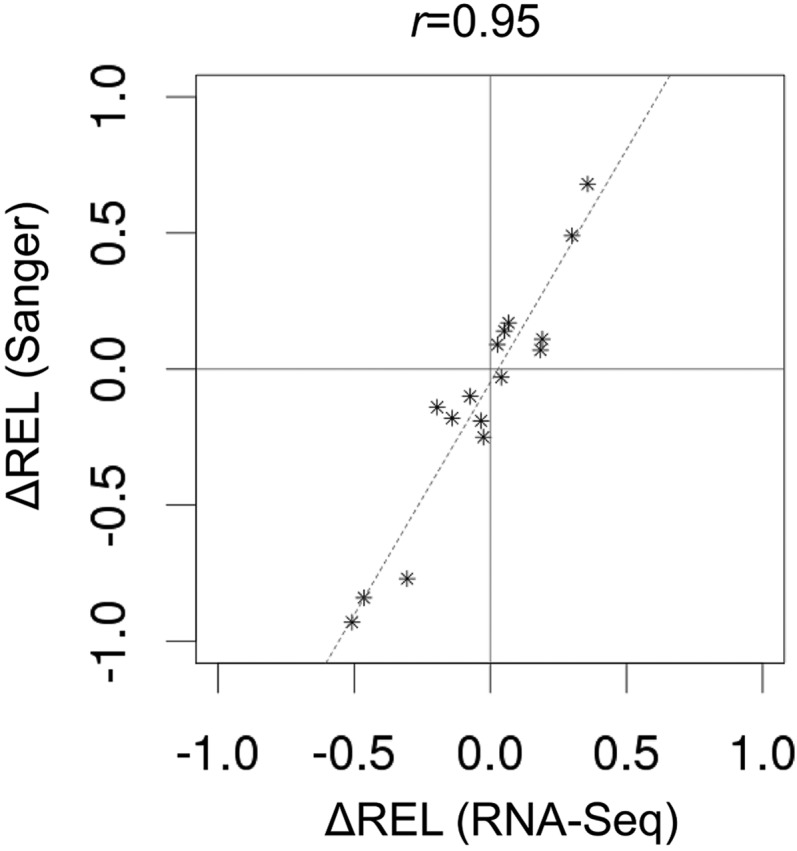
Comparison of edQTN-associated changes in RNA editing estimates based on RNA-Seq data and Sanger sequencing. For each RNA editing site we chose two strains that carried two alternate alleles of edQTN. The direction and magnitude of change was calculated as the difference in editing levels between strains that carried a reference or alternative alleles (∆REL). Note that the RNA-Seq experiments were performed on F1-hybrids, while the Sanger sequencing experiments were performed on homozygous inbred lines (see Materials and Methods). The Pearson’s correlation coefficient (r = 0.95, P < 10−8), and line of best fit are indicated (dashed line). Correlation between RNA editing estimates (REL) based on two methods is shown in Figure S1.
