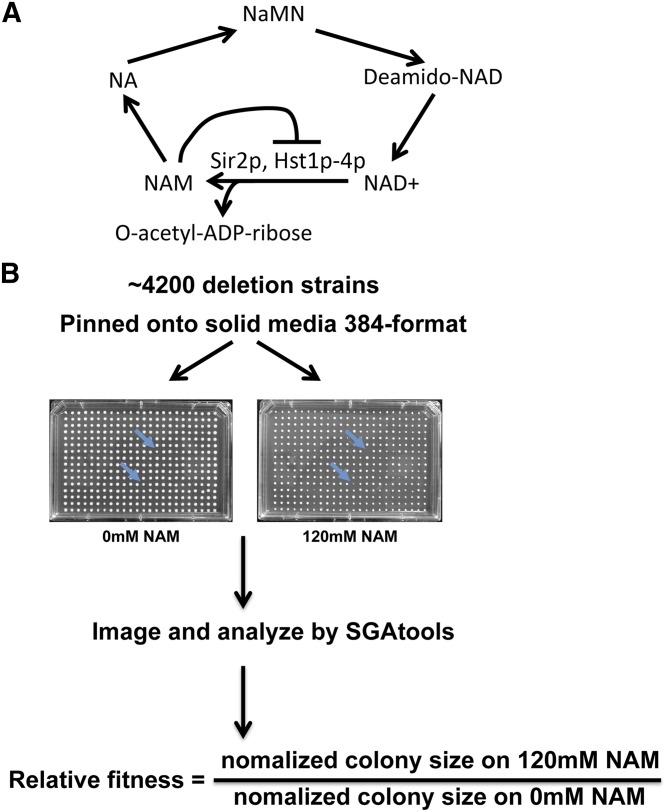
Figure 1.
A genome-wide screen for identifying deletion mutants sensitive to NAM. (A) Diagram showing the general pathway utilized by yeast to generate nicotinamide adenine dinucleotide (NAD+). Sirtuins use NAD+ as a cofactor and generate nicotinamide (NAM) as well as 2′O-acetyl ADP-ribose during a single deacetylation event. NAM can be converted to nicotinic acid (NA), which in turn is used to generate nicotinic acid mononucleotide (NaMN). Next, NaMN is converted to Deamido-NAD, and in turn NAD+ is regenerated. (B) Approach used to screen and score deletion mutants that are sensitive to NAM. A collection of ∼4200 yeast deletion mutants were arrayed in 384-format on YPD agar containing 0 or 120 mM NAM. Arrows indicate examples of the fitness defect observed in NAM sensitive mutants. Relative fitness was determined from normalized colony sizes obtained by analysis of plate images using SGAtools.