Figure 4. Refinement of the Hsp90 binding region on Nox5.
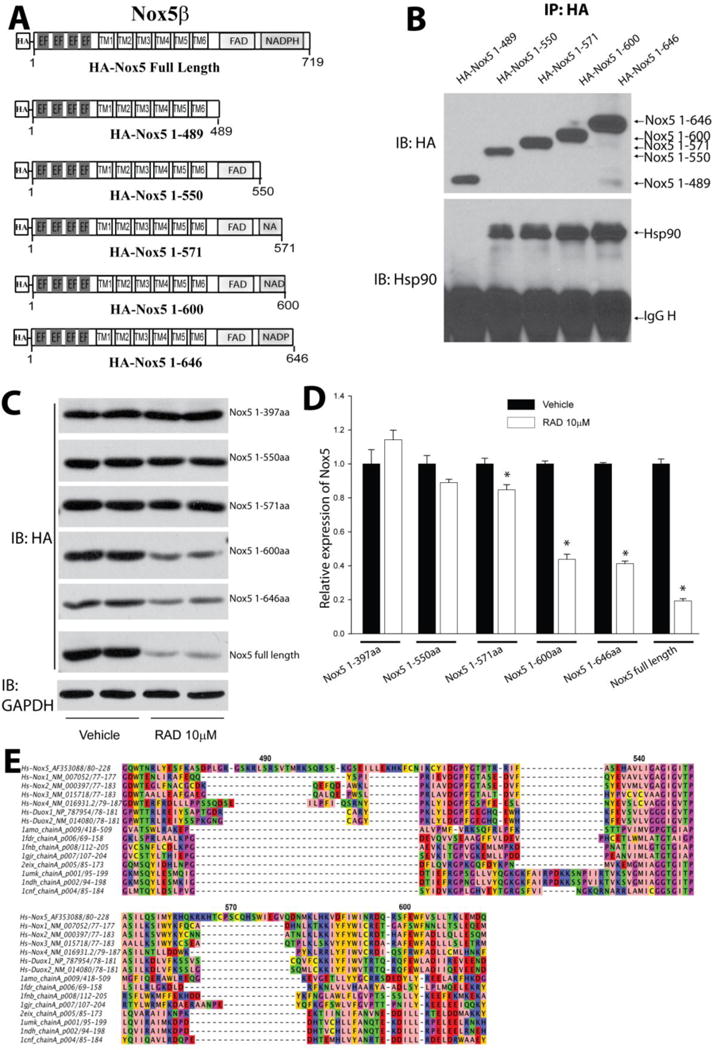
(A) Schematic of various truncation constructs of HA-Nox5β. (B) COS-7 cells were transfected with cDNAs encoding truncated Nox5 enzymes and the degree of Hsp90 binding determined by immunoprecipitation and western blot. (C) COS-7 cells expressing different Nox5 truncations were treated with indicated concentration of RAD for 24h, and expression levels determined by western blot. (D) Relative densitometry of Nox5 truncations vs. GAPDH expression. Results are representative of at least 3 separate experiments, presented as means ± S.E., *p<0.05 versus Vehicle. (E) Sequence analysis of HSP target regions on Nox5. A structurally informed multiple sequence alignment was constructed from dehydrogenase domains of Nox homologs and several crystallized dehydrogenases. A portion of the resulting alignment is shown, with color coding according to amino acid physicochemical properties using the Jalview Zappo scheme, and with Nox5 sequence numbering indicated. Sequence name (protein gi number)from top to bottom: Human Nox5alpha (gi 14211137), Human Nox1 isoform 1 (gi 148536873), Human Nox2 (gi 6996021), Human Nox3 (gi 11136626), Human Nox 4 (gi 8393843), Human Duox1 (gi 28872751), Human Duox2 (gi 132566532), Rat NADPH-cytochrome P450 Reductase (gi 3318958), E. coli Flavodoxin Reductase (gi 157831052), Spinach Ferredoxin Reductase (gi 157831108), Anabaena Ferredoxin NADP+ Reductase (gi 21730170), Physarum polycephalum Cytochrome B5 Reductase (gi 146387239), Human NADH cytochrome B5 Reductase (gi 56554196), Pig Cytochrome b5 Reductase (gi 999817), Corn Nitrate Reductase (gi 1064998).
