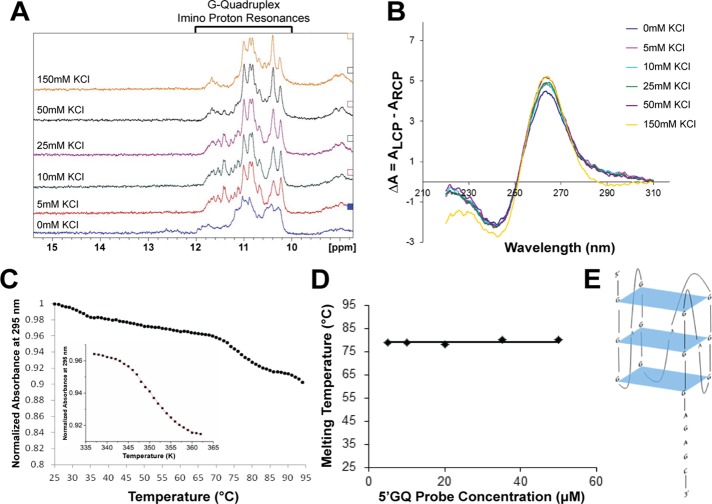
FIGURE 6:
The Gap-43 5′-UTR GQ sequence folds into a stable, parallel, intramolecular GQ Structure. (A) 1D 1H NMR spectroscopy with the Gap-43 5′GQ RNA probe revealed that imino proton resonances are present in the 10–12 ppm region even in the absence of KCl. (B) CD spectra of the Gap-43 5′GQ RNA probe in the presence of increasing KCl concentrations were acquired, and the results fitted the signature parallel GQ curve (negative peak at ∼240 nm and a positive peak at ∼265 nm). (C) UV spectroscopy thermal denaturation of the Gap-43 5′GQ RNA probe. Inset: fit of the main hypochromic transition present in the UV thermal denaturation profile of the Gap-43 5′GQ RNA probe with Eq. 4 (Materials and Methods) from which the following thermodynamic parameters were determined: ΔHo = −64.3 ± 0.1 kcal/mol, ΔSo = −183.2 ± 0.1 cal/mol K and ΔGo = −9.6 ± 0.1 kcal/mol. (D) Gap-43 5′GQ RNA probe melting temperature at 5 mM KCl as a function of the RNA concentration. (E) Arrangement of the predicted GQ structure within the Gap-43 5′GQ RNA probe. QGRS Mapper software was used for prediction (Kikin et al., 2006).