ABSTRACT
Silica is deposited in and around the spore coat layer of Bacillus cereus, and enhances the spore's acid resistance. Several peptides and proteins, including diatom silaffin and silacidin peptides, are involved in eukaryotic silica biomineralization (biosilicification). Homologous sequence search revealed a silacidin-like sequence in the C-terminal region of CotB1, a spore coat protein of B. cereus. The negatively charged silacidin-like sequence is followed by a positively charged arginine-rich sequence of 14 amino acids, which is remarkably similar to the silaffins. These sequences impart a zwitterionic character to the C terminus of CotB1. Interestingly, the cotB1 gene appears to form a bicistronic operon with its paralog, cotB2, the product of which, however, lacks the C-terminal zwitterionic sequence. A ΔcotB1B2 mutant strain grew as fast and formed spores at the same rate as wild-type bacteria but did not show biosilicification. Complementation analysis showed that CotB1, but neither CotB2 nor C-terminally truncated mutants of CotB1, could restore the biosilicification activity in the ΔcotB1B2 mutant, suggesting that the C-terminal zwitterionic sequence of CotB1 is essential for the process. We found that the kinetics of CotB1 expression, as well as its localization, correlated well with the time course of biosilicification and the location of the deposited silica. To our knowledge, this is the first report of a protein directly involved in prokaryotic biosilicification.
IMPORTANCE Biosilicification is the process by which organisms incorporate soluble silicate in the form of insoluble silica. Although the mechanisms underlying eukaryotic biosilicification have been intensively investigated, prokaryotic biosilicification was not studied until recently. We previously demonstrated that biosilicification occurs in Bacillus cereus and its close relatives, and that silica is deposited in and around a spore coat layer as a protective coating against acid. The present study reveals that a B. cereus spore coat protein, CotB1, which carried a C-terminal zwitterionic sequence, is essential for biosilicification. Our results provide the first insight into mechanisms required for biosilicification in prokaryotes.
INTRODUCTION
Silicon (Si), the second most abundant element in the Earth's crust, is an important mineral for living organisms. It acts as the main component of structural skeletons of diatoms, radiolarians, and siliceous sponges (1). Si is also utilized by some plants (e.g., rice and cucumber) to protect against biotic and abiotic stresses (2). The presence of Si in bacterial spores was first noted several decades ago (3, 4). Until recently, however, there have been no reports on the precise localization of Si and its biological function in prokaryotes. In a recent study, we demonstrated that the Bacillus cereus group, a very homogenous cluster of six species, and its close relatives accumulate Si in and around their spore coat layer and that the Si-containing layer enhances acid resistance of the spores (5). In Si-accumulating organisms, Si is taken up from the environment as soluble silicate (Si[OH]4), a biologically available form of Si in nature, which is then polymerized and accumulated as insoluble silica (SiO2). Such silica biomineralization (biosilicification) in eukaryotes is under genetic control, which in turn implies the existence of specific gene products guiding biosilicification. However, no proteins or peptides involved in prokaryotic biosilicification have been identified thus far.
The best-known example of biosilicification occurs in diatoms, which are eukaryotic unicellular algae that possess cell walls consisting of amorphous silica with a species-specific micro- and nano-patterning (6). To date, several peptides and proteins involved in diatom biosilicification have been identified, including silaffins, silacidins, and silaffin-like cingulins (7–9). The silaffins are short peptides that undergo extensive posttranslational modifications such as phosphorylation and polyamine conjugation. Due to a zwitterionic character imparted by negative charges of the phosphate groups and positive charges of the polyamines, silaffins easily self-assemble into supramolecular aggregates, which serve as the templates for silica formation (7, 10–12). The negatively charged silacidin peptides also form supramolecular aggregates with positively charged long-chain polyamines, and act as the templates for silica formation (8, 13). These observations indicate the importance of combining positive and negative charges in order to direct biosilicification.
Spores of Bacillus species are highly resilient dormant cell types that can withstand extremes of temperature, radiation, and chemical assault (14). These spore properties are attributable to the physical and chemical composition of the structures that encase the spore (15, 16). The outermost portion of Bacillus spores consists of cortex, spore coat, and, in some species, exosporium. The spore coat, where silica is deposited in B. cereus, is composed of more than 50 proteins (17). Silica accumulation in and around the B. cereus spore coat strongly suggests that spore coat proteins play an important role in biosilicification. We report here that one of the spore coat proteins, CotB1, carries a negatively charged silacidin-like sequence on its C terminus, followed by a positively charged arginine-rich sequence. We demonstrate that the zwitterionic C terminus of CotB1 plays an essential role in prokaryotic biosilicification.
MATERIALS AND METHODS
In silico screening for silica-forming proteins.
A database of B. cereus spore coat protein sequences (as reported by Henriques et al. [17]) was constructed with the sequences obtained from the NCBI website (http://www.ncbi.nlm.nih.gov/). Previously characterized silica-forming peptides (silaffin-1A1, -1A2, and -1B of Cylindrotheca fusiformis [7, 18] and silacidins A, B, and C of Thalassiosira pseudonana [9]) were used as query sequences. Searching for spore coat proteins with significant similarity to the silica-forming peptides was conducted using the BLAST program obtained from the NCBI FTP server (ftp://ftp.ncbi.nih.gov/blast/) with an E value of 10 as a threshold.
Bacterial strains and growth conditions.
B. cereus strain NBRC 15305, which corresponds to strain ATCC 14579 (19), was obtained from the NITE Biological Resource Center (NBRC; Chiba, Japan) and used as the wild-type strain in the present study. The wild-type and mutant strains of B. cereus were routinely grown at 28°C in LB medium (20). Sporulation was induced at 28°C by nutrient exhaustion in mR2A medium (an R2A medium [21] supplemented with 0.6 mM CaCl2, 0.03 mM MnCl2, 0.05 mM ZnCl2, and 0.05 mM FeSO4). We note that the concentrations of CaCl2 and MnCl2 in the medium are three times higher than in our previous report (5) in order to induce efficient sporulation for B. cereus. Spore development was monitored by bright-field microscopy, and the timing of entry into sporulation was defined as the end of the exponential growth phase (22). Escherichia coli JM109 was used as a host for cloning and was grown at 37°C in 2× YT medium (20). When necessary, spectinomycin (250 μg ml−1 for B. cereus), erythromycin (10 μg ml−1 for B. cereus), and carbenicillin (50 μg ml−1 for E. coli) were added to the medium.
Mutant construction.
The cotB1B2 deletion mutant was constructed by using the allelic replacement method described by Arnaud et al. (23). DNA fragments (∼1 kb each) corresponding to the up- and downstream regions of cotB1B2 were amplified from the chromosomal DNA of B. cereus using the primer pairs cotB1-F1/cotB1-R1 and cotB2-F1/cotB2-R1 (nucleotide sequences are given in Table S1 in the supplemental material) and then treated with BglII and SalI, respectively. A spectinomycin resistance (Spr) cassette was amplified from pIC333 (24) with the primers spc-F1 and spc-R1 and was digested with BglII and SalI. These three fragments were ligated by T4 DNA ligase, and the resultant fragment was amplified by PCR with the primers cotB1-F1 and cotB2-R1. The PCR fragment was digested with MluI and inserted into the MluI site of the pMAD vector (23), which was obtained from the Pasteur Institute (Paris, France), yielding pMADΔcotB1B2 (Table 1). The plasmid was then introduced into the B. cereus wild-type strain by electroporation (25), and the transformants were selected at 30°C on LB agar plates with 250 μg ml−1 spectinomycin and 50 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Integration and excision of pMADΔcotB1B2 were performed as described by Arnaud et al. (23) with slight modifications: the double-crossover deletion mutants were screened by blue/white screening on LB agar plates without spectinomycin, because they showed unexpectedly low spectinomycin resistance when the Spr cassette was inserted into the cotB1B2 locus. The insufficient resistance can be ascribed to the incomplete promoter sequence of the Spr cassette (26) and the upstream cotB1B2 promoter, which is not active in vegetative cells (see Results and Discussion). These weak promoters could not support sufficient expression of the Spr cassette, although the cassette works appropriately when inserted into the tetB locus of the wild-type strain (23). Disruption of cotB1B2 was confirmed by PCR amplification with the primer pairs cotB1-F2/spc-R2 and cotB2-R2/spc-F2.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pIC333 | Source of an Spr cassette | 24 |
| pMAD | Thermosensitive shuttle vector for allelic replacement; Ampr in E. coli, Emr in B. cereus | 23 |
| pMADΔcotB1B2 | pMAD carrying up- and downstream regions of cotB1B2 with an inserted Spr cassette | This study |
| pMADΔcotG | pMAD carrying up- and downstream regions of cotG with an inserted Spr cassette | This study |
| pHT304 | Shuttle vector; Ampr in E. coli, Emr in B. cereus | 29 |
| pHT-cotB1B2 | pHT304 carrying cotB1 and cotB2 with the promoter region of cotB1B2 | This study |
| pHT-cotB1 | pHT304 carrying cotB1 with the promoter region of cotB1B2 | This study |
| pHT-cotB2 | pHT304 carrying cotB2 with the promoter region of cotB1B2 | This study |
| pHT-cotB1-141 | pHT-cotB1 lacking silacidin-like and arginine-rich sequences | This study |
| pHT-cotB1-157 | pHT-cotB1 lacking arginine-rich sequence | This study |
| pHT-cotB1-142/157 | pHT-cotB1 lacking silacidin-like sequence | This study |
| pGFPuv | Source of gfp gene | Clontech Laboratories |
| pHT-gfp-cotB1 | pHT304 carrying gfp-fused cotB1 | This study |
| pHT-gfp | pHT304 carrying gfp | This study |
Ampr, ampicillin resistance; Emr, erythromycin resistance; Spr, spectinomycin resistance.
The cotG deletion mutant was constructed in a similar way by replacing the primers cotB1-F1, cotB1-R1, cotB2-F1, and cotB2-R1 with cotG-F1, cotG-R1, cotG-F2, and cotG-R2, respectively, to yield pMADΔcotG. Disruption of cotG was confirmed by PCR amplification with the primer pairs cotG-F3/spc-R2 and cotG-R3/spc-F2.
Extraction of spore coat proteins.
Spores were prepared by growing B. cereus strains in mR2A medium supplemented with 100 μg ml−1 silicate at 28°C for 48 h and were harvested by centrifugation. The pellets were sonicated in buffer containing 25 mM Tris-HCl (pH 7.5), 1 mM EDTA, 15% glycerol, 0.1 M NaCl, and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) to disrupt residual mother cells, and then the spores were separated from the suspension by centrifugation as reported by Isticato et al. (27). Spore coat proteins were extracted from the spores with sodium dodecyl sulfate (SDS) gel-loading buffer (20) containing 50 mM Tris-HCl (pH 6.8), 100 mM dithiothreitol, 10% glycerol, 2% SDS, and 0.1% bromophenol blue at 100°C for 10 min. The extracts were immediately fractionated on 12.5% Bis-Tris SDS-PAGE gels with 3-(N-morpholino)propanesulfonic acid running buffer (28) and stained with Coomassie brilliant blue R-250.
Silicate uptake assay.
B. cereus cells grown overnight with shaking at 28°C in R2A medium were inoculated (5%) into mR2A medium supplemented with 100 μg ml−1 silicate. The cultures were incubated at 28°C with shaking. Samples taken from the cultures at 12-h intervals were centrifuged, and then the silicate concentrations in the supernatants were measured by using a silicate test kit (Merck, Darmstadt, Germany) according to the manufacturer's instructions.
Acid resistance assay.
Sporulation of B. cereus strains was induced in mR2A medium with or without 100 μg ml−1 silicate as described above. After the cultures were kept standing at 4°C for 12 h, the spores were collected by centrifugation at 8,100 × g for 10 min and washed twice with cold sterile distilled water. The purified spores were suspended in distilled water to an optical density of 1.0 at 600 nm. Spores were centrifuged and resuspended in equal volumes of 0.2 N HCl. At the designated times, samples were diluted with cold water and spread on R2A agar plates. Spore viability was determined by counting the colonies after a 24-h incubation at 28°C.
Plasmid construction and transformation.
For complementation tests, a DNA fragment containing the cotB1B2 genes with their putative promoter and terminator regions was amplified by PCR from the chromosomal DNA of B. cereus using the primers cotB1-F3 and cotB2-R3 (see Table S1 in the supplemental material). The PCR fragment was digested with BamHI and EcoRI and then inserted into the same sites of plasmid pHT304 (29), yielding pHT-cotB1B2. The plasmids carrying either cotB1 or cotB2 gene were amplified by inverse PCR using pHT-cotB1B2 as a template with the primer pairs cotB1-F4/cotB1-R2 and cotB2-F2/cotB2-R4, respectively. The amplified DNA fragments were self-ligated to yield pHT-cotB1 and pHT-cotB2, respectively. To construct the plasmids carrying truncated mutants of cotB1, another round of inverse PCR was performed using pHT-cotB1 as a template with the primer pairs cotB1-F5/cotB1-R3, cotB1-F6/cotB1-R3, and cotB1-F6/cotB1-R4. The amplified DNA fragments were self-ligated to yield pHT-cotB1-141, pHT-cotB1-157, and pHT-cotB1-142/157, respectively. These plasmids were introduced into the ΔcotB1B2 strain by electroporation as described above, and then the transformants were selected at 28°C on LB agar plates with 10 μg ml−1 erythromycin. To construct a plasmid expressing green fluorescent protein (GFP)-fused CotB1 under the control of the promoter of cotB1B2, the vector backbone was amplified by inverse PCR using pHT-cotB1 as a template with the primers cotB2-F2 and cotB1-R5. A GFP-encoding DNA fragment was amplified by PCR from pGFPuv (Clontech Laboratories, Mountain View, CA) using the phosphorylated primers gfp-F1 and gfp-R1. These PCR products were ligated by T4 DNA ligase, yielding pHT-gfp-cotB1. As a control, plasmid pHT-gfp, which expresses GFP under the control of the promoter of cotB1B2, was prepared by the same procedures using the primer pairs cotB2-F2/cotB1-R2 and gfp-F1/gfp-R2. These plasmids were introduced into the wild-type strain by electroporation as described above.
Microscopy.
Sporulating cells and spores grown in mR2A medium supplemented with silicate were harvested at various time points and observed under a BX51 fluorescence microscope equipped with an UPlanApo ×100/1.35 objective lens and a U-MNIBA3 filter (Olympus, Tokyo, Japan). Images were captured using a DP72 cooled charge-coupled device camera (Olympus). For transmission electron microscopy (TEM), spores were prepared without silicate as described above. Ultrathin sections of the spores were prepared and stained as described previously (5) except that spores were initially fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) containing 0.1% MgSO4 for 4 h at 4°C, followed by postfixation with 1% OsO4 in 0.2 M cacodylate buffer (pH 7.2) for 10 h at 4°C. The ultrathin sections were observed under a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan) operating at an accelerating voltage of 80 kV.
RESULTS AND DISCUSSION
Silacidin-like sequence in the C-terminal region of CotB1.
To identify the proteins involved in the biosilicification of B. cereus spores, we performed a local BLAST search of spore coat proteins using sequences of previously characterized silica-forming peptides (diatom silaffins and silacidins) as queries. Among the 53 spore coat proteins that have been identified in B. cereus to date (17), a C-terminal region of CotB1 (GenBank accession number AAP07429.1) showed significant similarity to silacidin A (50% identity over 16 amino acids, E value 8.5) (Fig. 1, boxed). Silacidin A is a highly acidic peptide of 28 amino acids that is isolated from cell walls of the diatom T. pseudonana (9). Interestingly, a gene encoding a CotB1 paralog, namely, CotB2 (GenBank accession number AAP07430.1), is located in tandem downstream of the cotB1 gene on the B. cereus genome (see Fig. S1 in the supplemental material). The cotB1 and cotB2 genes are separated by only 20 bp. The cotB1 gene is preceded by a putative σK promoter (30), which is activated in the mother cell during the late stage of sporulation, whereas no canonical promoter sequence is found in the 500-bp sequence preceding the cotB2 open reading frame. These facts imply that the cotB1 and cotB2 genes are present in a single transcriptional unit. Although CotB1 and CotB2 are highly similar to each other (49% identity and 66% similarity over 131 amino acids), CotB1 is longer than CotB2 (171 versus 149 amino acids) and contains a characteristic C-terminal extension (Fig. 1). The extension overlaps the silacidin A-like sequence identified by the local BLAST search (residues 142 to 157) (Fig. 1, boxed), which is rich in serine, aspartate, and glutamate residues, as is the case for silacidin A. The downstream region (residues 158 to 171) (Fig. 1, underlined) is rich in serine and positively charged arginine residues. Such properties are common to diatom silaffins, although the majority of positively charged residues in silaffins are lysines (e.g., silaffin R5 [SSKKSGSYSGSKGSKRRIL]) (7). The presence of the negatively charged silacidin-like sequence and positively charged arginine-rich sequence would impart a zwitterionic character to the C-terminal extension of CotB1.
FIG 1.
Pairwise alignment of B. cereus CotB1 and CotB2 and primary structure of T. pseudonana silacidin A. Alignments of CotB1 with CotB2 (A) and with silacidin A (B) were performed using the CLUSTAL W program at GenomeNet (http://www.genome.jp/) with default parameters. Black-shaded and gray-shaded amino acid residues indicate identity and similarity, respectively. The boxed sequence of CotB1 (CGESDESDDSKESSDN) was identified by a local BLAST search for sequences with significant similarity to silacidin A (SGDSPPSDESEESEDS). The underlined sequence indicates the arginine-rich region of CotB1. The amino acid sequence of C. fusiformis silaffin-1A1 is given for comparison.
CotB1 is involved in the biosilicification of B. cereus spores.
In order to investigate whether CotB1 and/or CotB2 is involved in the biosilicification, we constructed a ΔcotB1B2 mutant strain. The mutant contains a chromosomal mutation that replaces the entire cotB1 and cotB2 genes with an Spr cassette. We also constructed another mutant strain, a ΔcotG mutant, as a negative control, in which the Spr cassette replaces the gene encoding another spore coat protein Cotγ (31) (formerly known as an exosporium protein ExsB [32]; however, recent studies showed that most of this protein is located in the spore coat [31, 33]). These mutant strains grew as fast and formed spores at the same rate as the wild type in mR2A medium with 100 μg ml−1 silicate (data not shown), suggesting that the cotB1B2 and cotG disruption did not affect vegetative growth or spore formation. Biosilicification of the spores was evaluated by measuring the decrease in silicate concentration in the culture supernatant during incubation, because our previous study indicated that most of the silicate removed from the culture supernatant is deposited as silica in the spores (5). During the early stationary phase (24 to 36 h after inoculation), the silicate concentration in the culture supernatants of the wild type and ΔcotG mutant decreased by about 75 μg ml−1, while that in the ΔcotB1B2 culture supernatant decreased by only about 25 μg ml−1 (Fig. 2A), indicating that one or both of the cotB1B2 genes are involved in biosilicification.
FIG 2.
Silicate uptake and spore acid resistance of the B. cereus wild type and ΔcotB1B2 and ΔcotG mutants. (A) Silicate concentrations in the culture supernatant of the wild-type (●), ΔcotB1B2 (○), and ΔcotG (▲) strains were measured at the indicated times after inoculation into mR2A medium supplemented with 100 μg ml−1 silicate. Microscopic observation showed that engulfed forespores formed during the 12 to 24 h period and that mature spores were gradually released from the mother cells after 36 h. (B) Viability of wild-type (circles) and ΔcotB1B2 (triangles) spores in 0.2 N HCl. Spores were produced in the presence (closed symbols) or absence (open symbols) of silicate. The data represent the means and standard deviations of the results of at least three independent experiments.
We previously reported that the biosilicification of the spore coat increased spore viability under acidic conditions (5). To investigate whether cotB1B2 disruption affects acid resistance, we purified spores from the wild-type and mutant strains, which were produced in mR2A medium with or without silicate, and treated them with 0.2 N HCl. Consistent with our previous results (5), the wild-type spores prepared in the presence of silicate showed higher survival rates than those prepared in the absence of silicate (Fig. 2B). This was also the case for the ΔcotG mutant (data not shown). In contrast, and as expected, the presence of silicate did not affect the survival rates of the ΔcotB1B2 spores (Fig. 2B). The ΔcotB1B2 spores prepared with or without silicate showed survival rates similar to those of the wild-type spores prepared without silicate (Fig. 2B). These data indicate that biosilicification-associated acid resistance was significantly reduced in the ΔcotB1B2 spores.
In order to exclude the possibility that the decrease in the silicate uptake and acid resistance in the ΔcotB1B2 mutant was caused by incomplete coat assembly, we examined wild-type and ΔcotB1B2 spores following thin-section TEM and found no measurable differences in spore coat structure (see Fig. S2 in the supplemental material). Consistent with this, SDS-PAGE analysis of spore coat proteins from the ΔcotB1B2 mutant showed specific loss of a protein with a molecular mass consistent with that of CotB1 (19.3 kDa) in the ΔcotB1B2 spores (Fig. 3, arrowhead). The absence of CotB2 could not be unequivocally identified on the SDS gel because of the overlap with other highly expressed protein bands near the expected molecular mass of CotB2 (16.8 kDa). These results indicate that the disruption of cotB1B2 genes did not measurably disrupt the overall spore coat assembly, as reported for several other spore coat proteins (34–36). Therefore, the decrease in acid resistance in the ΔcotB1B2 mutant is not due to incomplete coat assembly.
FIG 3.
SDS-PAGE analysis (12.5%) of the spore coat proteins. Sporulation of the B. cereus wild type (WT) and ΔcotB1B2 mutant was induced in mR2A medium supplemented with 100 μg ml−1 silicate for 48 h. The arrowhead indicates the position corresponding to the expected size of CotB1 (19.3 kDa).
To perform gene complementation analysis, we constructed plasmids carrying cotB1, cotB2, or both of these genes with the native promoter region of cotB1B2, designated pHT-cotB1, pHT-cotB2, and pHT-cotB1B2, respectively. Wild-type levels of biosilicification were restored by the introduction of either pHT-cotB1 or pHT-cotB1B2 into the ΔcotB1B2 mutant, whereas this was not the case for pHT-cotB2 (Fig. 4A). The reduction in acid resistance caused by cotB1B2 disruption was also restored by the introduction of pHT-cotB1 and pHT-cotB1B2 but not pHT-cotB2 (data not shown). These results clearly indicate that only CotB1 is involved in the biosilicification.
FIG 4.
Complementation analysis of ΔcotB1B2. (A) Silicate concentrations in the culture supernatant of the B. cereus wild-type strain carrying pHT304 (●), ΔcotB1B2 carrying pHT304 (○), pHT-cotB1B2 (■), pHT-cotB1 (□), or pHT-cotB2 (▲) were measured at the indicated times after inoculation into mR2A medium supplemented with 100 μg ml−1 silicate. (B) The same experiment was performed for the wild-type strain carrying pHT304 (●), ΔcotB1B2 carrying pHT304 (○), pHT-cotB1 (■), pHT-cotB1-141 (□), pHT-cotB1-157 (▲), or pHT-cotB1-142/157 (△). The data represent the means and standard deviations of the results of at least three independent experiments.
A C-terminal zwitterionic sequence is essential for biosilicification.
As noted above, CotB1 (but not CotB2) has a C-terminal extension consisting of a negatively charged silacidin-like region (residues 142 to 157) and a positively charged arginine-rich region (residues 158 to 171) (Fig. 1). To investigate whether the zwitterionic extension is essential for biosilicification, we constructed three plasmids encoding C-terminally truncated mutants of CotB1: pHT-cotB1-141 encodes CotB1 lacking residues 142 to 171, pHT-cotB1-157 encodes CotB1 lacking residues 158 to 171, and pHT-cotB1-142/157 encodes CotB1 lacking the residues between 142 and 157. Introduction of any of these mutant plasmids into the ΔcotB1B2 did not restore the biosilicification (Fig. 4B), although comparable amounts of the C-terminally truncated CotB1 proteins were expressed and integrated in the spores (see Fig. S3 in the supplemental material). These results strongly suggest that both the silacidin-like and arginine-rich sequences, which would impart a zwitterionic character to the C terminus of CotB1, are essential for biosilicification.
Expression and localization of CotB1.
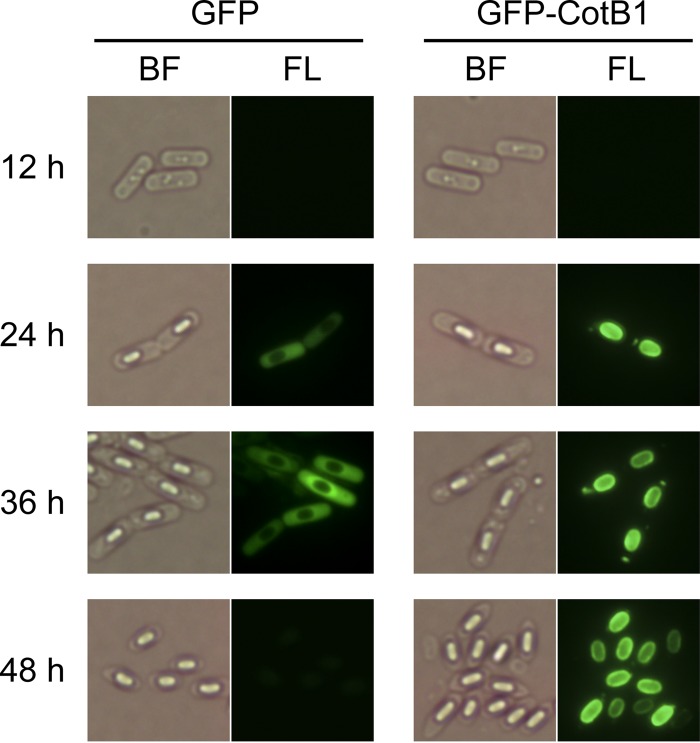
To further examine the relationship between CotB1 and biosilicification, we monitored the time course of expression and subcellular localization of CotB1 during sporulation. First, we constructed a plasmid containing the cotB1B2 promoter region followed by a chimeric gene, gfp-cotB1, that encodes CotB1 with an N-terminal GFP (pHT-gfp-cotB1), and then we introduced it into the ΔcotB1B2 mutant. However, the levels of biosilicification were not completely restored in the transformant (data not shown), implying that the presence of the fused GFP attenuates CotB1 function. Therefore, we introduced pHT-gfp-cotB1 into the wild-type strain, producing a merodiploid strain that contains gfp-cotB1 fusion on the plasmid in addition to the native cotB1 chromosomal locus. The transformant grew and sporulated normally in mR2A medium, and its levels of biosilicification were the same as those of the wild-type strain (data not shown). In the merodiploid strain, no fluorescence signals were detected under fluorescence microscopy at 12 h after inoculation, which is approximately 2 h after entry into sporulation (Fig. 5). At 24 h after inoculation, when the silicate uptake started and the engulfed forespores could be observed by bright-field microscopy, green fluorescence was distributed around the entire forespores (Fig. 5). This expression time course agrees well with the presence of a putative σK promoter (30) upstream of the cotB1 gene, which is activated in the mother cell during the late stage of sporulation (see Fig. S1 in the supplemental material). Subsequently, GFP fluorescence was consistently associated with the developing spores, and finally formed a complete ring around the mature spores. This fluorescence localization pattern is essentially the same as those of previously reported GFP-fused spore proteins located in the exosporium and/or spore coat (37–39), where silica is deposited in B. cereus (5). In contrast, in the wild-type strain carrying pHT-gfp that contains only the gfp gene instead of gfp-cotB1, GFP fluorescence was dispersed throughout the mother cell cytoplasm and did not localize on the spore. Taken together, the timing of CotB1 expression and its localization agree well with the time course of biosilicification and the location of the deposited silica, supporting the direct involvement of this protein in silica formation.
FIG 5.
Bright-field and fluorescence microscopic analysis of GFP-CotB1 during sporulation. The B. cereus wild-type strains carrying pHT-gfp-cotB1 (GFP-CotB1) or pHT-gfp (GFP) were grown in mR2A medium supplemented with 100 μg ml−1 silicate. Samples were taken at the indicated times after inoculation and observed by bright-field (BF) and fluorescence (FL) microscopy.
CotB1 may be the founding member of a subset of prokaryotic proteins that stimulate biosilicification.
As described above, the C-terminal zwitterionic sequence of CotB1 is similar to diatom silica-forming peptides, silaffins and silacidins (Fig. 1), and is essential for biosilicification in vivo (Fig. 4B). In contrast, no proteins homologous to sponge silicatein α, which is involved in biosilicification in siliceous sponges (40, 41), are found in the B. cereus genome. These facts suggest that B. cereus and diatoms share similar molecular mechanisms for silica formation, although the very limited overlapping lengths between CotB1 and silaffins/silacidins prevent further investigation into the evolutionary relationships among them. Scheffel et al. proposed that preassembled protein-based templates are general components of the cellular machinery for silica morphogenesis in diatoms (8). Our findings clearly indicate that this is also the case for bacteria, because B. cereus CotB1 is also a component of an insoluble supramolecular aggregate (the spore coat) that serves as a template for silica formation. To the best of our knowledge, this is the first report of an intracellular protein that stimulates biosilicification in prokaryotes.
The CotB1 homologs that contain the C-terminal zwitterionic sequence are common to biosilicifying B. cereus relatives (e.g., GenBank accession numbers AIF54819.1 [B. anthracis] and ADH05148.1 [B. thuringiensis]) but not to nonbiosilicifying Bacillus species (such as B. subtilis) or other organisms. These facts are consistent with our previous finding that all of the biosilicifying bacteria, which we isolated from paddy field soil, belong to or are closely related to the B. cereus group (5). Our findings indicate that CotB1 plays an important role in prokaryotic biosilicification. However, the presence of a CotB1 homolog is not sufficient for biosilicification. For example, biosilicification was not observed in the spores of B. mycoides (also a member of the B. cereus group) (5), although this bacterium has a CotB1 homolog with a zwitterionic sequence (GenBank accession number EEM01263.1). These findings clearly indicate the involvement of an additional factor(s) in the biosilicification of Bacillus spores. Such a factor(s) probably includes the presence of silicate transporters, because spore biosilicification is likely to require active transport of silicate into the mother cells. Since biosilicification occurs before the release of the spores (5), silicate needs to be taken up via transporters on the mother cell surface. The slight decrease in silicate concentration in the culture supernatant in the ΔcotB1B2 mutant (Fig. 2A) could be attributable to silicate uptake into the mother cells by the silicate transporters. Since the mutant cells cannot efficiently polymerize silicate, most of it is later released during mother cell lysis. In diatoms, silica deposition is tightly coupled to silicate transport, which is mediated by specific transporters (42, 43). Influx and efflux silicate transporters have been also identified in higher plants (44, 45). However, counterparts of the eukaryotic silicate transporters have not been identified in prokaryotes. Elucidation of the mechanism of silicate transport in B. cereus may help explain the differences in biosilicification among Bacillus species and could also provide new insights into the distribution of biosilicification in prokaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Didier Lereclus for the gift of the pHT304 plasmid. We also thank Kanae Koike for technical assistance in TEM analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00447-15.
REFERENCES
- 1.Lowenstam HA, Weiner S. 1989. On biomineralization. Oxford University Press, New York, NY. [Google Scholar]
- 2.Epstein E. 1999. Silicon. Annu Rev Plant Physiol Plant Mol Biol 50:641–664. doi: 10.1146/annurev.arplant.50.1.641. [DOI] [PubMed] [Google Scholar]
- 3.Jonhstone K, Ellar DJ, Appleton TC. 1980. Location of metal ions in Bacillus megaterium spores by high-resolution electron probe X-ray microanalysis. FEMS Microbiol Lett 7:97–101. doi: 10.1111/j.1574-6941.1980.tb01584.x. [DOI] [Google Scholar]
- 4.Stewart M, Somlyo AP, Somlyo AV, Shuman H, Lindsay JA, Murrell WG. 1980. Distribution of calcium and other elements in cryosectioned Bacillus cereus T spores, determined by high-resolution scanning electron probe X-ray microanalysis. J Bacteriol 143:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirota R, Hata Y, Ikeda T, Ishida T, Kuroda A. 2010. The silicon layer supports acid resistance of Bacillus cereus spores. J Bacteriol 192:111–116. doi: 10.1128/JB.00954-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Round FE, Crawford RM, Mann DG. 1990. The diatoms: biology and morphology of the genera. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 7.Kröger N, Deutzmann R, Sumper M. 1999. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 286:1129–1132. doi: 10.1126/science.286.5442.1129. [DOI] [PubMed] [Google Scholar]
- 8.Scheffel A, Poulsen N, Shian S, Kröger N. 2011. Nanopatterned protein microrings from a diatom that direct silica morphogenesis. Proc Natl Acad Sci U S A 108:3175–3180. doi: 10.1073/pnas.1012842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenzl S, Hett R, Richthammer P, Sumper M. 2008. Silacidins: highly acidic phosphopeptides from diatom shells assist in silica precipitation in vitro. Angew Chem Int Ed Engl 47:1729–1732. doi: 10.1002/anie.200704994. [DOI] [PubMed] [Google Scholar]
- 10.Kröger N, Lorenz S, Brunner E, Sumper M. 2002. Self-assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science 298:584–586. doi: 10.1126/science.1076221. [DOI] [PubMed] [Google Scholar]
- 11.Otzen D. 2012. The role of proteins in biosilicification. Scientifica 2012:867562. doi: 10.6064/2012/867562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumper M, Kröger N. 2004. Silica formation in diatoms: the function of long-chain polyamines and silaffins. J Mater Chem 14:2059–2065. doi: 10.1039/b401028k. [DOI] [Google Scholar]
- 13.Richthammer P, Börmel M, Brunner E, van Pée KH. 2011. Biomineralization in diatoms: the role of silacidins. ChemBioChem 12:1362–1366. doi: 10.1002/cbic.201000775. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev 64:548–572. doi: 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driks A. 1999. Bacillus subtilis spore coat. Microbiol Mol Biol Rev 63:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriques AO, Moran CP Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95–110. doi: 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- 17.Henriques AO, Moran CP Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 18.Kröger N, Deutzmann R, Sumper M. 2001. Silica-precipitating peptides from diatoms: the chemical structure of silaffin-1A from Cylindrotheca fusiformis. J Biol Chem 276:26066–26070. doi: 10.1074/jbc.M102093200. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 20.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 23.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinmetz M, Richter R. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J Bacteriol 176:1761–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turgeon N, Laflamme C, Ho J, Duchaine C. 2006. Elaboration of an electroporation protocol for Bacillus cereus ATCC 14579. J Microbiol Methods 67:543–548. doi: 10.1016/j.mimet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Murphy E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″)(9). Mol Gen Genet 200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- 27.Isticato R, Esposito G, Zilhão R, Nolasco S, Cangiano G, De Felice M, Henriques AO, Ricca E. 2004. Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J Bacteriol 186:1129–1135. doi: 10.1128/JB.186.4.1129-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Updyke TV, Engelhorn SC. 2000. System for pH-neutral stable electrophoresis gel. US patent 6,162,338.
- 29.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
- 30.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aronson A, Goodman B, Smith Z. 2014. The regulated synthesis of a Bacillus anthracis spore coat protein that affects spore surface properties. J Appl Microbiol 116:1241–1249. doi: 10.1111/jam.12452. [DOI] [PubMed] [Google Scholar]
- 32.Todd SJ, Moir AJG, Johnson MJ, Moir A. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J Bacteriol 185:3373–3378. doi: 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abhyankar W, Hossain AH, Djajasaputra A, Permpoonpattana P, Ter Beek A, Dekker HL, Cutting SM, Brul S, de Koning LJ, de Koster CG. 2013. In pursuit of protein targets: proteomic characterization of bacterial spore outer layers. J Proteome Res 12:4507–4521. doi: 10.1021/pr4005629. [DOI] [PubMed] [Google Scholar]
- 34.Cutting S, Zheng L, Losick R. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J Bacteriol 173:2915–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donovan W, Zheng L, Sandman K, Losick R. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol 196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 36.Zilhão R, Serrano M, Isticato R, Ricca E, Moran CP Jr, Henriques AO. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol 186:1110–1119. doi: 10.1128/JB.186.4.1110-1119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A. 2006. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol 59:487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- 38.Mallozzi M, Bozue J, Giorno R, Moody KS, Slack A, Cote C, Qiu D, Wang R, McKenney P, Lai EM, Maddock JR, Friedlander A, Welkos S, Eichenberger P, Driks A. 2008. Characterization of a Bacillus anthracis spore coat-surface protein that influences coat-surface morphology. FEMS Microbiol Lett 289:110–117. doi: 10.1111/j.1574-6968.2008.01380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson BM, Hsieh HY, Spreng KA, Stewart GC. 2011. The co-dependence of BxpB/ExsFA and BclA for proper incorporation into the exosporium of Bacillus anthracis. Mol Microbiol 79:799–813. doi: 10.1111/j.1365-2958.2010.07488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cha JN, Shimizu K, Zhou Y, Christiansen SC, Chmelka BF, Stucky GD, Morse DE. 1999. Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. Proc Natl Acad Sci U S A 96:361–365. doi: 10.1073/pnas.96.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu K, Cha J, Stucky GD, Morse DE. 1998. Silicatein α: cathepsin L-like protein in sponge biosilica. Proc Natl Acad Sci U S A 95:6234–6238. doi: 10.1073/pnas.95.11.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Jézéquel V, Hildebrand M, Brzezinski MA. 2000. Silicon metabolism in diatoms: implications for growth. J Phycol 36:821–840. doi: 10.1046/j.1529-8817.2000.00019.x. [DOI] [Google Scholar]
- 43.Thamatrakoln K, Hildebrand M. 2008. Silicon uptake in diatoms revisited: a model for saturable and nonsaturable uptake kinetics and the role of silicon transporters. Plant Physiol 146:1397–1407. doi: 10.1104/pp.107.107094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M. 2006. A silicon transporter in rice. Nature 440:688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- 45.Ma JF, Yamaji N, Mitani N, Tamai K, Konishi S, Fujiwara T, Katsuhara M, Yano M. 2007. An efflux transporter of silicon in rice. Nature 448:209–212. doi: 10.1038/nature05964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.