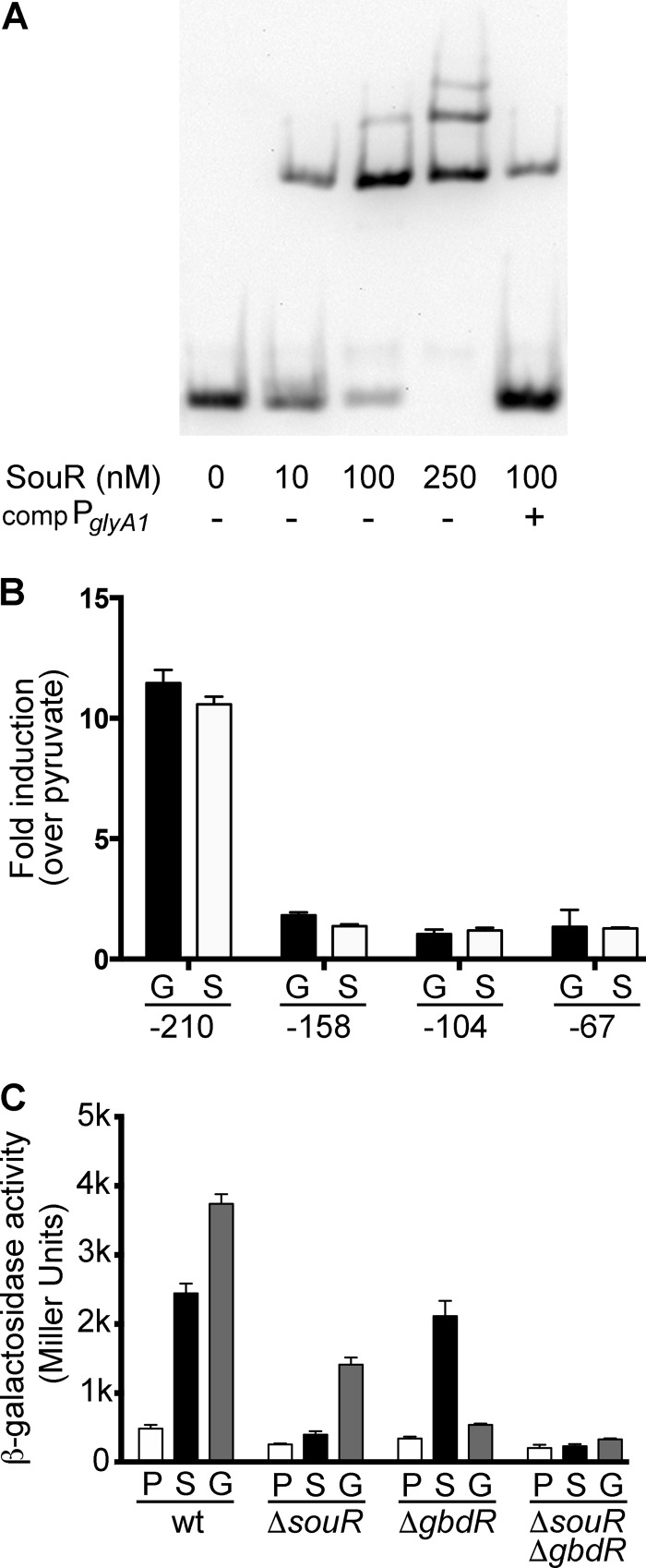
FIG 4.
SouR interaction with the PglyA1 promoter. (A) EMSA performed with MBP-SouR (SouR) and biotinylated PglyA1 probe. The data are representative of four independent experiments performed with two separately purified batches of MBP-SouR. The presence (+) or absence (−) of unlabeled competitor (comp) PglyA1 probe is noted below each lane. (B) Results from a β-galactosidase assay for promoter mapping to identify regions within PglyA1 required for souR- and gbdR-dependent induction. The ΔgbdR cells were exposed to 1 mM sarcosine (S), and ΔsouR cells were exposed to glycine betaine (G) and compared to controls with pyruvate. The size of each PglyA1 promoter construct is noted as the beginning position relative to the glyA1 translational start site. Fold induction was calculated as a multiple of the pyruvate condition for each strain. (C) Results from a β-galactosidase assay for the −210 PglyA1 promoter in wild-type, ΔsouR, ΔgbdR, and ΔsouR ΔgbdR strains. Cells were induced as for panel B. The data shown are representative of three biological replicates, and the error bars represent standard deviations.