ABSTRACT
Gram-negative bacteria express a number of sophisticated secretion systems to transport virulence factors across the cell envelope, including the type II secretion (T2S) system. Genes for the T2S components GspC through GspN and PilD are conserved among isolates of Acinetobacter baumannii, an increasingly common nosocomial pathogen that is developing multidrug resistance at an alarming rate. In contrast to most species, however, the T2S genes are dispersed throughout the genome rather than linked into one or two operons. Despite this unique genetic organization, we show here that the A. baumannii T2S system is functional. Deletion of gspD or gspE in A. baumannii ATCC 17978 results in loss of secretion of LipA, a lipase that breaks down long-chain fatty acids. Due to a lack of extracellular lipase, the gspD mutant, the gspE mutant, and a lipA deletion strain are incapable of growth on long-chain fatty acids as a sole source of carbon, while their growth characteristics are indistinguishable from those of the wild-type strain in nutrient-rich broth. Genetic inactivation of the T2S system and its substrate, LipA, also has a negative impact on in vivo fitness in a neutropenic murine model for bacteremia. Both the gspD and lipA mutants are outcompeted by the wild-type strain as judged by their reduced numbers in spleen and liver following intravenous coinoculation. Collectively, our findings suggest that the T2S system plays a hitherto-unrecognized role in in vivo survival of A. baumannii by transporting a lipase that may contribute to fatty acid metabolism.
IMPORTANCE Infections by multidrug-resistant Acinetobacter baumannii are a growing health concern worldwide, underscoring the need for a better understanding of the molecular mechanisms by which this pathogen causes disease. In this study, we demonstrated that A. baumannii expresses a functional type II secretion (T2S) system that is responsible for secretion of LipA, an extracellular lipase required for utilization of exogenously added lipids. The T2S system and the secreted lipase support in vivo colonization and thus contribute to the pathogenic potential of A. baumannii.
INTRODUCTION
Acinetobacter baumannii, an increasingly common nosocomial Gram-negative pathogen, is responsible for a wide range of infections, including pneumonia, urinary tract infections, bacteremia, meningitis, and skin and wound infections (1–4). Immunocompromised and severely ill patients in the intensive care unit, individuals with extensive wounds or invasive devices, and those who are undergoing or have recently undergone antibiotic regimens are particularly susceptible to A. baumannii infections (5–7). Ventilator-associated pneumonia and bloodstream infections are the most severe, resulting in 25% to 35% mortality rates (8, 9).
The pathogenic success of A. baumannii is likely multifactorial, but its ability to persist on dry surfaces, form biofilms, resist complement-mediated killing, and survive antibiotic treatment are of importance (1, 10–13). The escalating frequency of multidrug-resistant (MDR) strains of A. baumannii is of particular concern. In the past 10 years, there has been an alarming 60% increase in the number of MDR clinical isolates reported (http://www.cddep.org). An important and clinically relevant aspect of bacterial infections is the ability of bacteria to grow as a biofilm (14). These matrix-encased, multilayer bacterial communities are exceptionally resistant to antibiotic treatment and are prone to spreading antibiotic resistance through horizontal gene transfer (15, 16). Clinical isolates that form biofilms survive for long periods of time on dry surfaces and are able to colonize common hospital equipment, such as ventilator tubes (17). Several factors have been shown to be necessary for abiotic biofilm formation, including a pilus assembly system, the OmpA outer membrane protein, and capsular polysaccharide, which is also protective against complement-mediated killing (13, 18–20).
While research has focused on the mechanisms of antibiotic resistance and biofilm formation and the epidemiology of A. baumannii, our understanding of A. baumannii pathogenesis is lagging and little is known about the contribution of secreted proteins to A. baumannii survival and propagation during infection. Sequencing of several A. baumannii genomes has revealed that A. baumannii contains genes for a variety of transport systems, including the assembly and translocation system for type IV pilus and type VI and type IV secretion systems. The type IV pilus supports the twitching motility of A. baumannii (21) but may also contribute to adhesion, colonization, biofilm formation, and transformation like type IV pili in other Gram-negative pathogens, while the type VI secretion system is utilized for bacterial competition and the type IV secretion system may be required for virulence (22, 23). In addition, A. baumannii possesses genes for a type II secretion (T2S) system (24, 25). Bacteria that use the T2S system are typically environmental bacteria; however, they also include pathogens such as Vibrio cholerae, enterotoxigenic Escherichia coli, Pseudomonas aeruginosa, and Legionella pneumophila (26–29). The T2S system mediates the secretion of toxins and hydrolytic enzymes, including proteases, lipases, lipoproteins, and enzymes that break down complex carbohydrates, and has been found to be required for in vivo survival and virulence (30–39). Following inner membrane translocation via the Sec or TAT pathways, T2S substrates engage with the T2S system for transport across the outer membrane. This multiprotein secretion system is encoded by 12 to 16 general secretion pathway (gsp) genes (40–42). With rare exceptions, mutations in any of the core gsp genes, gspC through gspM and pilD, prevent extracellular secretion (41).
The gsp genes are scattered throughout the A. baumannii genome instead of being organized into one or two operons (27). Due to their unusual arrangement, it was unclear whether the gsp genes of A. baumannii encode a functional secretion system. Here, we demonstrate that the T2S system in A. baumannii is functional and identify a lipase as one of its secreted substrates. We show that the extracellular lipase, LipA, and the T2S system that transports this enzyme across the outer membrane are required for utilization of exogenously added lipids and support colonization of A. baumannii in a murine model of bacteremia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains listed in Table 1 were cultured in Luria-Bertani (LB) broth or on LB agar at 37°C. Carbenicillin (100 μg/ml) was used for plasmid maintenance.
TABLE 1.
Plasmids and bacterial strains
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Plasmids | ||
| pK18mobsacB | Suicide vector containing sacB (Kmr) | 77 |
| pCVD442 | Suicide vector containing sacB (Apr) | 78 |
| pMMB67EH | Low-copy-number, IPTG-inducible vector (Apr) | 79 |
| pgspD | pMMB67EH-gspD | This study |
| pgspE1 | pMMB67EH-gspE1 | This study |
| pgspE2 | pMMB67EH-gspE2 | This study |
| pgspN | pMMB67EH-gspN | This study |
| plipA | pMMB67EH-lipA | This study |
| plipBA | pMMB67EH-lipBA | This study |
| Strains | ||
| E. coli MC1061 | F− lac mutant; K-12 laboratory strain | 80 |
| E. coli MM294/pRK2013 | Helper strain for conjugation | 81 |
| E. coli SY327λpir | λpir lysogen; permits replication of pCVD442 | 82 |
| A. baumannii AYE | Clinical strain | 83 |
| A. baumannii AB0057 | Clinical strain | 84 |
| A. baumannii AB5075 | Clinical strain | 85 |
| A. baumannii 17978 | Wild type for T2S | ATCC |
| A. baumannii ΔgspD | Replacement of gspD with aph-3 (Kmr) | This study |
| A. baumannii ΔgspE1 | Replacement of gspE1 with aph-3 (Kmr) | This study |
| A. baumannii ΔgspE2 | Replacement of gspE2 with aph-3 (Kmr) | This study |
| A. baumannii ΔgspN | Replacement of gspN with aph-3 (Kmr) | This study |
| A. baumannii ΔlipA | Replacement of lipA with aph-3 (Kmr) | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance.
Construction of ΔlipA, ΔgspD, ΔgspE1, ΔgspE2, and ΔgspN strains.
Chromosomal DNA isolated from the wild-type (WT) A. baumannii ATCC 17978 strain was used as the template for PCR. PCRs were carried out with Phusion DNA polymerase. Primers were synthesized by IDT Technologies.
To generate the ΔgspD strain, we used the primers indicated in Table 2 to amplify and clone 500 bp of DNA upstream and downstream of the gspD gene as well as the aph-3 kanamycin cassette into pCVD442. Transconjugates in which pCVD442 had recombined into the A. baumannii genome were selected on LB agar containing carbenicillin and chloramphenicol. To select for the second recombination event, individual colonies were cultured overnight in LB broth, diluted, cultured to the late log phase, and spread on LB agar containing 3% sucrose. Sucrose and kanamycin-resistant, carbenicillin-sensitive isolates were screened for loss of growth on lipid agar. Strains that were kanamycin resistant and carbenicillin sensitive were designated ΔgspD mutants. The deletion was verified by PCR. All other gene deletion strains (the ΔlipA, ΔgspE1, ΔgspE2, and ΔgspN mutants) were constructed in a similar manner using the appropriate primers shown in Table 2.
TABLE 2.
Primers used for plasmid construction
| Primer | Sequence (5′–3′) | Plasmid construct(s) generated |
|---|---|---|
| KanUp | CCGGAATTGCCAGCTGGG | ΔgspD, ΔgspE1, ΔgspE2, ΔgspN, and ΔlipA strains |
| KanDown | TTCAGAAGAACTCGTCAAG | ΔgspD, ΔgspE1, ΔgspE2, ΔgspN, and ΔlipA strains |
| gspD1 | GGTTCAACTTCTTACCAATT | ΔgspD vector |
| gspD2 | CCCAGCTGGCAATTCCGGTAAAGCCATAACTCGCGA | ΔgspD vector |
| gspD3 | CTTGACGAGTTCTTCTGAAGCGCCGTAGTAGCATGTTA | ΔgspD vector |
| gspD4 | CGGTGCGGGTTTTGGCACAG | ΔgspD vector |
| gspD5 | CGCGGATCCGTCACCATAAGAGTTAGGAA | pgspD |
| gspD6 | CGCGCATGCCATAACATGCTACTACGGCGCTG | pgspD |
| gspE1-1 | GTTAAACAGACTTCACGCTG | ΔgspE1 vector |
| gspE1-2 | CCCAGCTGGCAATTCCGGGTTTCAGG | ΔgspE1 vector |
| gspE1-3 | CTTGACGAGTTCTTCTGAAAGTGAAG | ΔgspE1 vector |
| gspE1-4 | GGTGCTGTAACTAACCCAG | ΔgspE1 vector |
| gspE1-5 | GACGAGCTCCTCATCATTATAAATTGT | pgspE1 |
| gspE1-6 | GACGTCGACGCATTTTTTATATCTTAC | pgspE1 |
| gspE2-1 | GAGCCTAGTCTCTTTTTTAA | ΔgspE2 vector |
| gspE2-2 | CTCTTGCGACATGACTTGCTTTTTCTTCATTCAGCC | ΔgspE2 vector |
| gspE2-3 | CTTGACGAGTTCTTCTGACCATTAAATTAAATTTTT | ΔgspE2 vector |
| gspE2-4 | CTACACGTTTTAAAGGCTTATAATC | ΔgspE2 vector |
| gspE2-5 | GACGTCGACATAGATGAGGTGAATCTT | pgspE2 |
| gspE2-6 | GACGAATTCATATATTGGGGAAAACAC | pgspE2 |
| gspN1 | GTTGAACAGCTTCTAGAATTTGGCG | ΔgspN vector |
| gspN2 | CCCAGCTGGCAATTCCGGCTTTTTCTTCATTCAGCC | ΔgspN vector |
| gspN3 | CTTGACGAGTTCTTCTGAGGTGGTAACTAATGAAAG | ΔgspN vector |
| gspN4 | GCTCTGTAGGTTGAGACGGTGTAGC | ΔgspN vector |
| gspN5 | CACGAATTCCATGTTGGTAAGGCTGAATG | pgspN |
| gspN6 | GCGAAGCTTCCATACTTTCATTAGTT | pgspN |
| lipA1 | AAGCTTGTCGACTTACACACACGTAC | ΔlipA vector |
| lipA2 | GTTGCATGCCGGTTAAAACCCGCCAT | ΔlipA vector |
| lipA3 | TTAGAGCTCCAAGGATTATAAGCTTT | ΔlipA vector |
| lipA4 | TTACCCGGGTTGATATGCGCTTTA | ΔlipA vector |
| lipA5 | GAGGAATTCAGTAAAAAATGAAAAGG | plipA |
| lipA6 | GAGGTCGACTAAAGCGTAAGCTTATA | plipA |
| lipBA1 | CAACGAGCTCAAACTTAAGGAAGATA | plipBA |
| lipBA2 | GAGGTCGACTAAAGCGTAAGCTTATA | plipBA |
Construction of plipBA, plipA, pgspE1, pgspE2, pgspN, and pgspD plasmids.
The lipA and lipB genes were amplified from chromosomal DNA using the appropriate primers shown in Table 2. The product was ligated into a low-copy-number, broad-host-range vector, pMMB67EH, to make plipBA. This broad-host-range expression vector has been used in many Gram-negative species, including P. aeruginosa and V. cholerae (43), and is stably maintained in A. baumannii. The construct was verified by sequencing and conjugated from the E. coli MC1061 strain into WT and mutant A. baumannii strains. The plasmids overexpressing lipA, gspD, gspE1, gspE2, and gspN were constructed in the same manner.
Lipid agar.
Selective agar was utilized to detect extracellular lipase activity. The medium was prepared as described previously (44) with modified minimal medium (47.8 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, 18.7 mM NH4Cl2, 0.1% Tween 20, 0.2 mM CaCl2), 40 μg/ml of neutral red, and 0.5% filtered-sterilized olive oil.
Lipase assay.
The strains were grown in LB broth supplemented with IPTG (isopropyl-β-d-thiogalactopyranoside) at a 50 μM final concentration to induce the expression of plasmid-encoded LipA. Following 16 h of growth, supernatants and cells were separated by centrifugation at 3,500 rpm for 10 min. A spectrophotometric assay was used to measure lipase activity by incubating culture supernatant with 0.9 mM 4-nitrophenyl myristate–80 mM Tris-HCl (pH 8.0)–0.15% Triton X-100 buffer at 37°C and measuring the release of 4-nitrophenol at 415 nm over time. All assays were performed in triplicate, and means and standard deviations of the results are presented.
SDS-PAGE and immunoblotting.
Culture supernatants were concentrated by precipitation utilizing pyrogallol red-molybdate-methanol as described previously (30). The samples were normalized to equivalent optical densities at 600 nm (OD600) and subjected to SDS-PAGE and immunoblot analysis using antibodies raised against A. calcoaceticus LipA (1:100) (45) and goat anti-rabbit IgG–horseradish peroxidase (HRP). Immunoblots were imaged using a Typhoon Trio (Amersham Biosciences).
Serine hydrolase probe.
Overnight cultures of WT/plipBA and ΔgspD/plipBA strains were grown in LB broth–50 μM IPTG at 37°C. Supernatants and cells were separated by centrifugation at 3,500 rpm for 10 min. ActiveX FP serine hydrolase probe (0.5 μl) (Thermo Scientific) was added to 25 μl of culture supernatants and incubated at room temperature for 60 min. Samples were matched by equivalent OD600 values, boiled in SDS sample buffer, subjected to SDS-PAGE on 4% to 12% Bis-Tris polyacrylamide gels (NuPAGE; Invitrogen), and visualized using a Typhoon Trio variable mode imager system and ImageQuant software.
In vitro competition assay.
WT and mutant A. baumannii strains were cultured separately overnight in LB broth at 37°C. Strains were diluted 1:100, and equivalent numbers of WT and mutant strains were pooled and cultured together at 37°C. At 0 (input), 8, and 24 h, aliquots of the mixed culture were diluted and plated on LB agar with or without kanamycin. CFU counts were determined after 24 h of incubation at 37°C. The competitive index (CI) was determined after 24 h as follows: CI = (mutant CFU/WT CFU)/(mutant input CFU/WT input CFU).
In vivo competition assay.
Eight-week-old female CBA/J mice (Jackson Laboratory) were injected intravenously with 150 μl and 100 μl of 20 mg/ml cyclophosphamide for 4 and 3 days, respectively, before the start of the experiment. Overnight cultures of WT, ΔlipA, and ΔgspD A. baumannii bacteria were diluted in phosphate-buffered saline (PBS). Inocula of 107 cells at 1:1 ratios of WT:ΔlipA or WT:ΔgspD bacteria were administered via tail-vein injection. After 24 h, mice were euthanized. Spleens and livers were removed and homogenized in PBS, and CFU counts were determined after plating on LB agar with and without kanamycin and 24 h of incubation at 37°C. CI values were calculated as described above.
Statistical tests.
A Wilcoxon signed-rank test was calculated for the CIs obtained for the in vitro and in vivo competition assays. A Student t test was performed for the lipase activity assay. Values were considered significant at P values of ≤0.05.
Ethics statement.
All mouse experiments were performed according to the protocol (PRO00005052) approved by the University Committee on Use and Care of Animals at the University of Michigan. This protocol is in complete compliance with the guidelines for humane use and care of laboratory animals mandated by the National Institutes of Health.
RESULTS
A. baumannii encodes a functional T2S system.
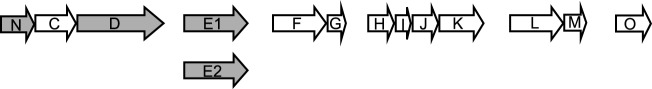
We analyzed the sequenced genome of several A. baumannii strains and performed homology searches with the T2S genes of V. cholerae and P. aeruginosa. We identified genes for each of the T2S components GspC through GspN and PilD (Fig. 1). In contrast to most species, these genes are scattered around the genome in six noncontiguous segments rather than being linked into one or two operons, perhaps as a result of genome plasticity and the remarkable ability of A. baumannii to acquire foreign DNA. To determine whether A. baumannii has a functional T2S system, we utilized A. baumannii ATCC 17978, a strain that was originally isolated from a 4-month-old child with fatal meningitis and that is amenable to genetic inactivation and plasmid-based complementation studies. Using allelic exchange, we inserted a kanamycin resistance gene cassette in place of gspD, gspE1, gspE2, and gspN. The gspD gene was chosen because in all studied T2S systems, GspD, the outer membrane pore that serves as the conduit through which proteins are transported, is absolutely essential for the T2S. GspE contributes energy for the secretion process by hydrolyzing ATP and is also indispensable for secretion. However, as A. baumannii carries two potential gspE genes, gspE1 and gspE2, we deleted both genes to resolve whether the gspE1 gene or the gspE2 gene or both are required for secretion in A. baumannii. In contrast to the roles of GspD and GspE, the role of GspN has not yet been determined and its gene is not present in every species with a functional T2S system. As gspN is localized in the same operon as gspD in A. baumannii, we wanted to determine whether it is also required for T2S in A. baumannii. To complement the deletion strains, we constructed expression vectors encoding the wild-type (WT) copy of each mutant gene and expressed them in trans.
FIG 1.
T2S genes in A. baumannii. Putative T2S components are encoded by gspN (N; A1S_0269), gspC (C; A1S_0270), gspD (D; A1S_0271), gspE1 (E1; A1S_0616), gspE2 (E2; A1S_2290), gspF (F; A1S_0369), gspG (G; A1S_0370), gspH (H; A1S_1562), gspI (I; A1S_1563), gspJ (J; A1S_1564), gspK (K; A1S_1565), gspL (L; A1S_2255), gspM (M; A1S_2254), and gspO (also named pilD) (O; A1S_0327). The genes shown in gray were deleted using allelic exchange technology.
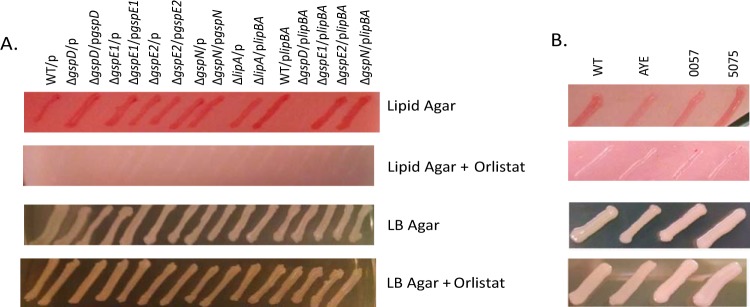
While the ΔgspD and ΔgspE1 mutants grew on LB agar as well as the WT, ΔgspE2, and ΔgspN strains, they were unable to grow on minimal agar with olive oil as the sole carbon source (Fig. 2A), a phenotype previously observed for T2S mutants of P. aeruginosa and V. cholerae due to their inability to secrete lipase (31, 44). Growth was restored when the ΔgspD and ΔgspE1 mutants were complemented with the appropriate expression plasmids (Fig. 2A).
FIG 2.
Growth on lipid agar requires LipA and a functional T2S system. Growth of strains on lipid agar, lipid agar with Orlistat, LB agar, and LB agar with Orlistat is shown from top to bottom. (A) Agar included carbenicillin and IPTG for plasmid maintenance and induction of expression of cloned genes. (B) Growth of reference strain ATCC 17978 (WT) and three clinical isolates.
Sequence analysis of A. baumannii ATCC 17978 identified genes with high homology to the genes of the P. aeruginosa and V. cholerae T2S substrate, LipA (Fig. 3), and its chaperone, LipB (46), suggesting that A. baumannii LipA may also be a T2S substrate that is capable of hydrolyzing lipids and generating nutrients for growth. To verify that LipA is the secreted substrate responsible for growth of A. baumannii on lipid agar, we constructed a lipA deletion strain by substituting the lipA gene for a gene encoding kanamycin resistance through homologous recombination. Similarly to the ΔgspD and ΔgspE1 mutants, the ΔlipA strain was unable to grow on the minimal lipid agar (Fig. 2A). Growth was restored when the lipA mutant was complemented with a plasmid encoding lipA and lipB. We also expressed the plasmid-encoded lipBA genes in the ΔgspD and ΔgspE1 deletion strain; however, despite overexpression of lipBA, no growth was observed on lipid agar (Fig. 2A). This suggests that the T2S system is active in A. baumannii and is responsible for the extracellular secretion of LipA.
FIG 3.
Sequence alignment of LipA from different species. The alignment of LipA from A. baumannii, V. cholerae, and P. aeruginosa is shown. The predicted N-terminal signal sequence is underlined. Yellow highlights indicate the catalytic residues.
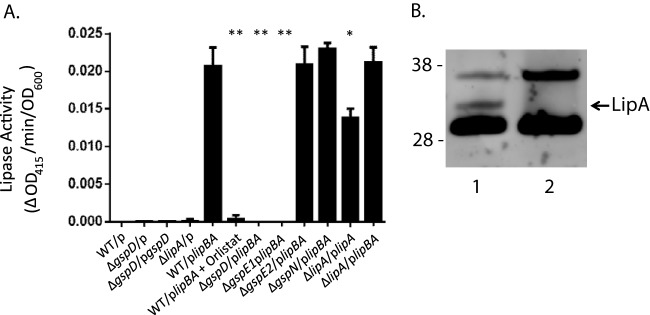
When A. baumannii is cultured in the absence of lipids in LB broth under standard laboratory conditions, the lipBA genes are likely not expressed. This is consistent with our inability to detect significant differences in lipase activity between culture supernatants of the WT, ΔgspD, ΔgspE1, ΔgspE2, ΔgspN, and ΔlipA strains (Fig. 4). Therefore, to quantitatively measure the level of LipA secretion, we induced expression of plasmid-encoded lipBA genes in the WT, ΔgspD, ΔgspE1, ΔgspE2, and ΔgspN strains. This resulted in detectable lipase activity in the WT/plipBA, ΔgspE2/plipBA, and ΔgspN/plipBA culture supernatants using 4-nitrophenyl myristate, while no activity was observed in the ΔgspD/plipBA and ΔgspE1/plipBA culture supernatants (Fig. 4A). In addition, immunoblotting with antibodies directed against A. calcoaceticus LipA resulted in the detection of extracellular LipA when overexpressed in WT A. baumannii but not in the ΔgspD mutant, providing further support for the idea of a requirement of a functional T2S system in extracellular secretion of LipA (Fig. 4B).
FIG 4.
Extracellular secretion of A. baumannii lipase is dependent on an intact T2S system. (A) Enzymatic activity of overnight culture supernatants against the lipase substrate 4-nitrophenyl myristate was measured as a change in absorbance at 415 nm per min and normalized to the absorbance of the culture at 600 nm. Bars show standard deviations from the means (*, P ≤ 0.05; **, P ≤ 0.001). (B) Concentrated stationary-phase WT/plipBA (lane 1) and ΔgspD/plipBA (lane 2) culture supernatants were subjected to SDS-PAGE and immunoblot analysis using LipA antibodies. Molecular mass markers are shown on the left. The position of LipA is indicated.
Characterization of the secreted substrate, LipA.
The A. baumannii T2S substrate, LipA, is 54% identical to the well-characterized P. aeruginosa LipA, which is a member of proteobacterial lipase homology group I (47). Lipases within this group require helper proteins (chaperones) for their proper folding (48, 49). This relationship appears to be true in A. baumannii as well. The ΔlipA/plipA strain, which overexpresses only the lipase, exhibited lower extracellular lipase activity than the ΔlipA/plipBA strain, where both the lipase and the chaperone are overexpressed (Fig. 4). This difference is likely due to a reduction in secretion of LipA, as it has been shown that proper folding of LipA is required for the outer membrane translocation of LipA via the T2S system in Pseudomonas glumae (49). Members of lipase homology group I have been classified as serine hydrolases with active sites containing the residues Ser-His-Asp/Glu (50, 51). Consistent with the overall high sequence homology, sequence alignments indicate that the A. baumannii LipA has the catalytic residues Ser-His-Asp (Fig. 3). We used the ActiveX serine hydrolase probe, which binds covalently to the serine nucleophile in the active site of serine hydrolases, to further examine the catalytic property of LipA. As LipA activity is not detected in the supernatant of WT A. baumannii, we incubated supernatants of WT/plipBA and ΔgspD/plipBA cultures induced with increasing amounts of IPTG with this probe. In the WT/plipBA samples, we observed a band increasing in intensity with increasing IPTG induction that corresponded to a protein of ∼32 kDa, the expected size of LipA (Fig. 5). In contrast, this band was not present in any of the ΔgspD/plipBA samples. Taken together with the sequence homology between LipA of A. baumannii and P. aeruginosa, these findings indicate that A. baumannii LipA is a secreted serine hydrolase belonging to lipase homology group I.
FIG 5.
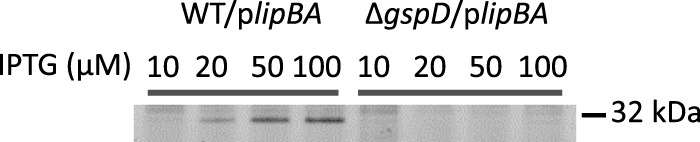
The ActiveX serine protease probe binds to LipA. Culture supernatants isolated from overnight WT/plipBA and ΔgspD/plipBA cultures induced with increasing concentrations (μM) of IPTG were incubated with the ActiveX serine protease probe for 1 h. Samples were subjected to SDS-PAGE and Typhoon image analysis as described in Materials and Methods. A molecular mass marker is shown on the right.
Orlistat is a semisynthetic, therapeutic drug prescribed for the treatment of obesity. It binds to the active site of human pancreatic lipase, thereby preventing the enzyme from breaking down dietary lipids in the intestine (52, 53). By incubating WT/plipBA culture supernatants with Orlistat, we learned that it also inhibits A. baumannii LipA (Fig. 4). Through its ability to inhibit LipA, Orlistat was therefore capable of inhibiting growth of A. baumannii when added to lipid agar but not when added to LB agar (Fig. 2). Similarly, the recently isolated clinical MDR strains of A. baumannii AYE, 0057, and 5075 were analyzed and shown to able to grow on lipid agar in the absence but not in the presence of Orlistat (Fig. 2B), suggesting that these MDR isolates also secrete lipases that can break down lipids and support fatty acid metabolism.
The T2S system and its secreted substrate, LipA, are required for A. baumannii colonization in a murine bacteremia model.
We utilized a modified murine bacteremia model originally developed for uropathogenic E. coli to determine if the T2S system and its substrate, LipA, are required for A. baumannii in vivo fitness (54). Due to the low virulence of A. baumannii ATCC 17978 in mice (55), we used an experimentally induced leukopenic mouse model (56), treating the mice with cyclophosphamide prior to infection. To diminish the potential of confounding factors between mice, we coinfected each mouse with equal numbers (1 × 107 CFU/ml) of either WT and ΔgspD bacteria or WT and ΔlipA bacteria. After 24 h, mice were euthanized and the spleens and livers were harvested and homogenized. Homogenates were plated on LB agar with and without kanamycin, CFU counts were determined, and the competitive index (CI) was calculated for each mutant strain as described in Materials and Methods (Fig. 6). A CI value below 1 indicates a colonization defect of the mutant relative to the WT strain. Both mutants were outcompeted by the WT strain and exhibited colonization defects in both the spleen and the liver. The reduction in colonization was not simply due to the presence of the kanamycin resistance cassette, as a strain with the Tn5 transposon containing the kanamycin resistance gene inserted into the ddc gene of A. baumannii showed no colonization defect in the spleen compared to the WT strain in a recent study using the same bacteremia model (56). In addition, the reduction in colonization of the ΔlipA and ΔgspD mutants is unique to the in vivo environment, as the ΔgspD and ΔlipA mutants grow as well as WT A. baumannii in LB broth (see Fig. S1 in the supplemental material) and in vitro competition experiments in LB broth did not show a difference between WT and ΔlipA or WT and ΔgspD strains (CI = 0.98 ± 0.76 and CI = 0.84 ± 0.58, respectively).
FIG 6.
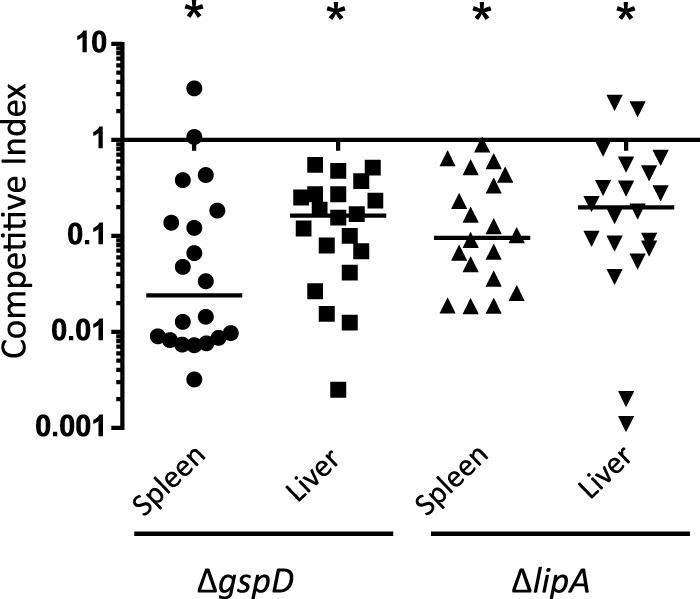
The gspD and lipA genes are required for A. baumannii fitness in a mouse model of bacteremia. Mice were coinoculated with equal numbers of either WT and ΔgspD or WT and ΔlipA strains. Mice were euthanized 24 h postinfection, the organs were harvested, and the CFU levels were determined for each strain. The competitive indexes were calculated as follows: (mutant CFU/WT CFU)/(mutant input CFU/WT input CFU). For all four competitions, P values = <0.001.
DISCUSSION
The T2S system controls the secretion of toxins and hydrolytic enzymes required for virulence in a variety of pathogens. Here, we present the results of the first study that identifies a functional T2S system in A. baumannii. Our study shows not only that A. baumannii possesses all the genes for a T2S system but also that there is a measurable reduction in extracellular LipA activity and loss of growth on lipid agar when gspD and gspE1, two of the core T2S genes, are deleted. In contrast, deletion of gspE2 has no effect on lipase secretion in A. baumannii. While GspE1 supports T2S and the homologous ATPase A1S_0329 likely provides energy for type IV pilus assembly, it is not clear what process GspE2 participates in, if any. Perhaps it affects T2S or type IV pilus biogenesis under very specific conditions. The finding that deletion of gspN had no effect on lipase secretion is consistent with its absence in many bacteria with functional T2S systems. In kind, the gspN homolog pulN is not required for pullulanase secretion via the T2S system in Klebsiella oxytoca (57). Similarly to GspE2, GspN may support T2S under only very specific growth conditions. Alternatively, it may substitute for GspC to support secretion of T2S substrates other than LipA (see below).
As mentioned earlier, the T2S genes are distributed throughout the genome in multiple operons in A. baumannii rather than in the one or two operons typical of other organisms encoding T2S systems. These smaller operons contain 1 to 4 gsp genes and, in some cases, may represent functional units that encode T2S components that directly interact. For example, GspC and GspD interact via the periplasmic domain of GspC and mutations that interfere with their interaction have a negative impact on secretion (58, 59). Interestingly, Xanthomonas campestris does not have a gspC gene; instead, it expresses GspN, which is required for secretion and has been shown to interact with GspD (60). Two proteins of the inner membrane platform, GspM and GspL, are encoded on another operon, and we have demonstrated in previous studies that these proteins stabilize each other within the cytoplasmic membrane and can be coimmunoprecipitated from V. cholerae (61). The minor pseudopilins GspI, GspJ and GspK interact and may form a subcomplex at the tip of the T2S pseudopilus (62), and they are encoded by the same operon that also contains the minor pseudopilin gspH gene. The genes for the inner membrane protein GspF and the major pseudopilin GspG are located in their own operon. Currently, there is no known interaction between GspF and GspG; however, the colocalization of their respective genes may suggest a potential functional interaction between these proteins in the T2S complex. Finally, while the cytoplasmic ATPase GspE forms a stable complex with GspL at the cytoplasmic membrane (63, 64), gspE1 and gspE2 are encoded separately from gspL on their own individual operons. The reason for this is not known, but, as discussed above, the data may suggest that GspE1 and GspE2 may be used under different conditions or for the secretion of different substrates. The unusual arrangement of the T2S genes presents an interesting avenue for further study.
While the exact role of LipA is currently unknown, it may be required for nutrient acquisition, breaking down lipids or long-chain fatty acids into shorter forms that are imported by the bacterium and used as sources of carbon and energy through β-oxidation (65). Besides being consumed as nutrients, fatty acids derived through lipid hydrolysis may be used as signaling molecules. Specifically, fatty acid signals are capable of restoring persister cells to a metabolically active state (66). The signaling molecule cis-2-dodecenoic acid contributes to virulence in cystic fibrosis Burkholderia cenocepacia infections (67) (68). While lipases may not have been considered virulence factors in the past, the P. aeruginosa LipA and LipC lipases contribute to motility, biofilm formation, pyoverdine production, and rhamnolipid production (69–71). Other lipases cleave host lipids to generate fatty acids that are integrated into the pathogens' own membranes (72). In addition, the extracellular esterase of group A Streptococcus reduces phagocyte recruitment by hydrolyzing platelet-activating factor (PAF) (73), and lipases produced and secreted by the opportunistic fungal pathogen Candida albicans support colonization and penetration of host cells (74, 75). Whether A. baumannii LipA is involved in nutrient acquisition or signaling or plays another important role in virulence will be explored in future studies.
The remarkable ability of A. baumannii to develop resistance to multiple antibiotics underscores the necessity for novel treatment approaches. Therapeutics that disarm A. baumannii or reduce its in vivo fitness are promising alternatives. One possibility is that of targeting LipA with a lipase inhibitor such as Orlistat. Alternatively, therapeutic targeting of secretion systems that transport multiple virulence factors to the exterior of the bacterium may be of particular interest, as their inactivation should have a greater impact than the targeting of individual virulence factors. It is quite possible that A. baumannii secretes additional proteins via the T2S besides LipA based on the following observations. First, the T2S system is active and supports secretion whether exogenous lipids are present or not, while lipA expression appears to require lipids/fatty acids (Fig. 2A and 4). Second, the T2S system is commonly required for the secretion of several proteins in other species. For example, proteomic analyses of culture supernatants of L. pneumophila, V. cholerae, and Burkholderia pseudomallei indicate that more than 20 different proteins may be dependent on the T2S system for extracellular release (30, 39, 76). Future work will focus on identifying additional T2S substrates and determining their role in A. baumannii pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Derek Samarian and Khalil Chedid for their assistance in the construction of strains and Klaas Hellingwerf for the LipA antibodies.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00622-15.
REFERENCES
- 1.Mak JK, Kim M-J, Pham J, Tapsall J, White PA. 2009. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother 63:47–54. [DOI] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi C, Malczynski M, Parker M, Scheetz MH. 2008. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J Clin Microbiol 46:1106–1109. doi: 10.1128/JCM.01877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck-Sagué CM, Jarvis WR, Brook JH, Culber DH, Potts A, Gay E, Shotts BW, Hill B, Anderson RL, Weinstein MP. 1990. Epidemic bacteremia due to Acinetobacter baumannii in five intensive care units. Am J Epidemiol 132:723–733. [DOI] [PubMed] [Google Scholar]
- 6.Playford EG, Craig JC, Iredell JR. 2007. Carbapenem-resistant Acinetobacter baumannii in intensive care unit patients: risk factors for acquisition, infection and their consequences. J Hosp Infect 65:204–211. doi: 10.1016/j.jhin.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Maragakis LL, Perl TM. 2008. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis 46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 8.Garnacho-Montero J, Ortiz-Leyba C, Jimenez-Jimenez F, Barrero-Almodovar A, Garcia-Garmendia J, Bernabeu-Wittell M, Gallego-Lara S, Madrazo-Osuna J. 2003. Treatment of multidrug-resistant Acinetobacter baumannii ventilator-associated pneumonia (VAP) with intravenous colistin: a comparison with imipenem-susceptible VAP. Clin Infect Dis 36:1111–1118. doi: 10.1086/374337. [DOI] [PubMed] [Google Scholar]
- 9.Falagas ME, Rafailidis PI, Kasiakou S, Hatzopoulou KP, Michalopoulos A. 2006. Effectiveness and nephrotoxicity of colistin monotherapy vs. colistin-meropenem combination therapy for multidrug-resistant Gram-negative bacterial infections. Clin Microb Infect 12:1227–1230. doi: 10.1111/j.1469-0691.2006.01559.x. [DOI] [PubMed] [Google Scholar]
- 10.Espinal P, Marti S, Vila J. 2012. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 80:56–60. doi: 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Wendt C, Dietze B, Dietz E, Ruden H. 1997. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 35:1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weernink A, Severin WPJ, Tjernberg I, Dijkshoorn L. 1995. Pillows, an unexpected source of Acinetobacter. J Hosp Infect 29:189–199. doi: 10.1016/0195-6701(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 13.Russo T, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 15.Mah T-F. 2012. Biofilm-specific antibiotic resistance. Future Microbiol 7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 16.Madsen JS, Burmolle M, Hansen LH, Sorenson SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 17.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomaras AP, Dorsey CW, Edelmann R, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 19.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litran T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RSJ. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carruthers MD, Nicholson PA, Tracy EN, Munson RSJ. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng T-T, Tyler BM, Setubal JC. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9(Suppl 1):S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020–1031. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cianciotto NP. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol 13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Sandkvist M. 2001. Type II secretion and pathogenesis. Infect Immun 69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filloux A. 2011. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol 2:155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rondelet A, Condemine G. 2013. Type II secretion: the substrates that won't go away. Res Microbiol 164:556–561. doi: 10.1016/j.resmic.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikora AE, Lybarger SR, Sandkvist M. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J Bacteriol 189:8484–8495. doi: 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwobi A, Heesemann J, Garcia E, Igwe E, Noelting C, Rakin A. 2003. Novel virulence-associated type II secretion system unique to high-pathogenicity Yersinia enterocolitica. Infect Immun 71:1872–1879. doi: 10.1128/IAI.71.4.1872-1879.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho TD, Davis BM, Ritchie JM, Waldor MK. 2008. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect Immun 76:1858–1865. doi: 10.1128/IAI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jyot J, Balloy V, Jouvion G, Verma A, Touqui L, Huerre M, Chignard M, Ramphal R. 2011. Type II secretion system of Pseudomonas aeruginosa: in vivo evidence of a significant role in death due to lung infection. J Infect Dis 203:1369–1377. doi: 10.1093/infdis/jir045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy-Simandle K, Stewart CR, Dao J, DebRoy S, Rossier O, Bryce PJ, Cianciotto NP. 2011. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect Immun 79:1984–1997. doi: 10.1128/IAI.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldi DL, Higginson EE, Hocking DM, Praszkier J, Cavaliere R, James CE, Bennett-Wood V, Azzopardi KI, Turnbull L, Lithgow T, Robins-Browne RM, Whitchurch CB, Tauschek M. 2012. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect Immun 80:2042–2052. doi: 10.1128/IAI.06160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandkvist M, Morales V, Bagdasarian M. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 123:81–86. doi: 10.1016/0378-1119(93)90543-C. [DOI] [PubMed] [Google Scholar]
- 38.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc Natl Acad Sci U S A 99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korotkov KV, Sandkvist M, Hol WGJ. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douzi B, Filloux A, Voulhoux R. 2012. On the path to uncover the bacterial type II secretion system. Philos Trans R Soc Lond B Biol Sci 367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow for direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
- 44.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn DN. 1998. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol Microbiol 27:221–233. [DOI] [PubMed] [Google Scholar]
- 45.Kok RG, van Thor JJ, Nugteren-Roodzant IM, Brouwer MBW, Egmond MR, Nudel CB, Vosman B, Hellingwerf KJ. 1995. Characterization of the extracellular lipase, LipA, of Acinetobacter calcoaceticus BD413 and sequence analysis of the cloned structural gene. Mol Microbiol 15:803–818. doi: 10.1111/j.1365-2958.1995.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 46.Ogierman MA, Fallarino A, Riess T, Williams SG, Attridge SR, Manning PA. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J Bacteriol 179:7072–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan ER, Leahy JG, Colwell RR. 1999. Cloning and sequence analysis of the lipase and lipase chaperone-encoding genes from Acinetobacter calcoaceticus RAG-1, and redefinition of a Proteobacterial lipase family and an analogous lipase chaperone family. Gene 230:277–285. doi: 10.1016/S0378-1119(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 48.Frenken LGJ, Bos JW, Visser C, Muller W, Tommassen J, Verrips CT. 1993. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol Microbiol 9:579–589. doi: 10.1111/j.1365-2958.1993.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 49.Frenken LGJ, de Groot A, Tommassen J, Verrips CT. 1993. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol Microbiol 9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 50.Jaeger K-E, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. 1994. Bacterial lipases. FEMS Microbiol Rev 15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 51.Snellman EA, Colwell RR. 2004. Acinetobacter lipases: molecular biology, biochemical properties and biotechnological potential. J Ind Microbiol Biotechnol 31:391–400. doi: 10.1007/s10295-004-0167-0. [DOI] [PubMed] [Google Scholar]
- 52.Heck AM, Yanovski JA, Calis KA. 2000. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy 20:270–279. doi: 10.1592/phco.20.4.270.34882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weibel EK, Hadvary P, Hochuli E, Kupfer E, Lengsfeld H. 1987. Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. I. Producing organism, fermentation, isolation and biological activity. J Antibiot (Tokyo) 40:1081–1085. [DOI] [PubMed] [Google Scholar]
- 54.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HLT. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eveillard M, Soltner C, Kempf M, Saint-Andre J-P, Lemarie C, Randrianarivelo C, Seifert H, Wolff M, Joly-Guillou M-L. 2010. The virulence variability of different Acinetobacter baumannii strains in experimental pneumonia. J Infect 60:154–161. doi: 10.1016/j.jinf.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, Zahdeh C, Mobley HLT. 2015. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere 1:00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Possot OM, Vignon G, Bomchil N, Ebel F, Pugsley AP. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J Bacteriol 182:2142–2152. doi: 10.1128/JB.182.8.2142-2152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korotkov KV, Johnson TL, Jobling MG, Pruneda J, Pardon E, Héroux A, Turley S, Steyaert J, Holmes RK, Sandkvist M, Hol WGJ. 2011. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog 7:e1002228. doi: 10.1371/journal.ppat.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korotkov KV, Krumm B, Bagdasarian M, Hol WGJ. 2006. Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae. J Mol Biol 363:311–321. doi: 10.1016/j.jmb.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 60.Lee H-M, Wang K-C, Liu Y-L, Yew H-Y, Chen L-Y, Leu W-M, Chen DC, Hu N-T. 2000. Association of the cytoplasmic membrane protein XpsN with the outer membrane protein XpsD in the type II protein secretion apparatus of Xanthomonas campestris pv. campestris. J Bacteriol 182:1549–1557. doi: 10.1128/JB.182.6.1549-1557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandkvist M, Hough LP, Bagdasarian MM, Bagdasarian M. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol 181:3129–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korotkov KV, Hol WGJ. 2008. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol 15:462–468. doi: 10.1038/nsmb.1426. [DOI] [PubMed] [Google Scholar]
- 63.Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J 14:1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WGJ. 2005. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J Mol Biol 348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 65.Houten SM, Wanders RJA. 2010. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J Inherit Metab Dis 33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marques CNH, Morozov A, Planzos P, Zelaya HM. 2014. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ Microbiol 80:6976–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. 2009. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol 191:5013–5019. doi: 10.1128/JB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang Y, Nguyen DT, Son MS, Hoang TT. 2008. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 b-oxidation operon. Microbiology 154:1584–1598. doi: 10.1099/mic.0.2008/018135-0. [DOI] [PubMed] [Google Scholar]
- 69.Funken H, Knapp A, Vasil ML, Wilhelm S, Jaeger K-E, Rosenau F. 2011. The lipase LipA (PA2862) but not LipC (PA4813) from Pseudomonas aeruginosa influences regulation of pyoverdine production and expression of the sigma factor PvdS. J Bacteriol 193:5858–5860. doi: 10.1128/JB.05765-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenau F, Isenhardt S, Gdynia A, Tielker D, Schmidt E, Tielen P, Schobert M, Jahn D, Wilhelm S, Jaeger K-E. 2010. Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa. FEMS Microbiol Lett 309:25–34. [DOI] [PubMed] [Google Scholar]
- 71.Tielen P, Kuhn H, Rosenau F, Jaeger K-E, Flemming H-C, Wingender J. 2013. Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa. BMC Microbiol 13:159–171. doi: 10.1186/1471-2180-13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pride AC, Herrera CM, Guan Z, Giles DK, Trent MS. 2013. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. mBio 4:e00305-13. doi: 10.1128/mBio.00305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu M, Zhu H, Li J, Garcia CC, Feng W, Kirpotina LN, Hilmer J, Tavares LP, Layton AW, Quinn MT, Bothner B, Teixeira MM, Lei B. 2012. Group A Streptococcus secreted esterase hydrolyzes platelet-activating factor to impede neutrophil recruitment and facilitate innate immune evasion. PLoS Pathog 8:e1002624. doi: 10.1371/journal.ppat.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu Y, Ibrahim AS, Fonzi W, Zhou X, Ramos CF, Ghannoum MA. 1997. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology 143:331–340. doi: 10.1099/00221287-143-2-331. [DOI] [PubMed] [Google Scholar]
- 75.Park M, Do E, Jung WH. 2013. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 41:67–72. doi: 10.5941/MYCO.2013.41.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burtnick MN, Brett PJ, DeShazer D. 2014. Proteomic analysis of the Burkholderia pseudomallei type II secretome reveals hydrolytic enzymes, novel proteins, and the deubiquitinase TssM. Infect Immun 82:3214–3226. doi: 10.1128/IAI.01739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 78.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using positive-selection suicide vector. Infect Immun 59:4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fürste JP, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 80.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol 138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 81.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller G, Feiss M. 1988. The bacteriophage λ cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol Gen Genet 212:157–165. doi: 10.1007/BF00322459. [DOI] [PubMed] [Google Scholar]
- 83.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie J-M, Raoult D, Médigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.