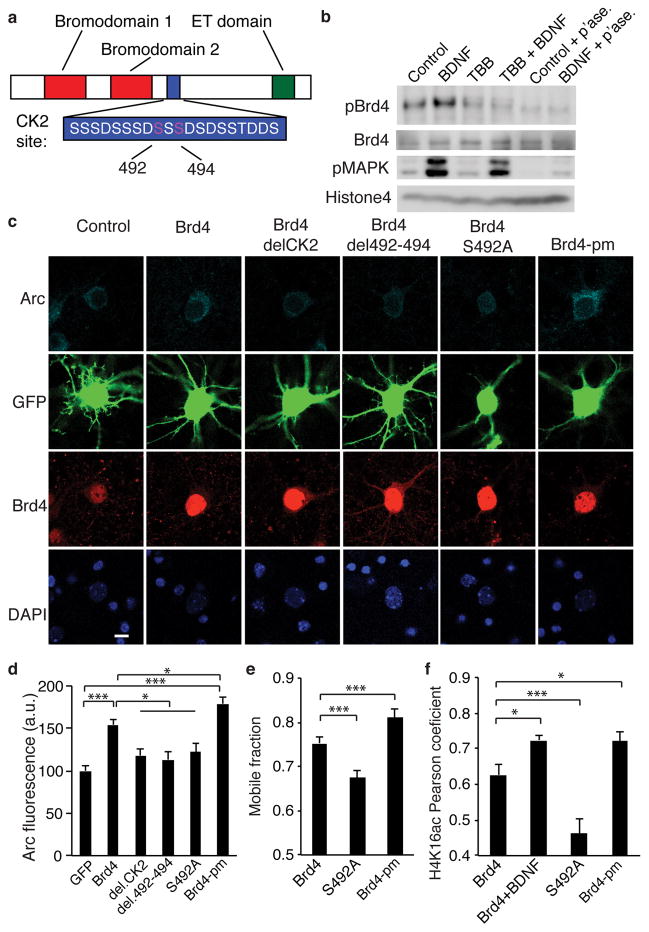
Figure 5. Phosphorylation of Brd4 is critical for its function.
(a) Model of Brd4 and the critical amino acids in the CK2 phosphorylation site. (b) Western blot for phosphorylated Brd4 shows an increase with BDNF but not after TBB pretreatment or phosphatase treatment of lysates. Representative of 3 biological replicates. (c, d) Staining (c) and quantification (d) for Arc and Brd4 in neurons transfected with GFP and Brd4 with deletions or mutations in the CK2 site (unpaired two-tailed t test, for GFP n = 68, for Brd4n = 61, for CK2 deletion n = 46, for deletion 492-494 n = 44, for S492A n = 54, for Brd4-pm n = 51 from 5 biological replicates, for GFP vs Brd4 P = 1.223E-8, t = 6.09, for GFP vs Brd4-pm P = 2.00E-13, t = 2.353, for Brd4 vs CK2 deletion P = 0.0011, t = 3.36, for Brd4 vs deletion 492-494 P = 0.00037, t = 3.689, for Brd4 vs S492A P = 0.0075, t = 2.724, for Brd4 vs Brd4-pm P = 0.0204, t = 2.353). (e) Quantification of the mobile fraction of FRAP performed on Brd4 with mutations in the CK2 domain (unpaired two-tailed t test, for Brd4 n = 57 neurons, for S492A n = 52 and for Brd4-pm n = 38 from 3 biological replicates, for Brd4 vs S492A P = 0.0001, t = 4.042, for Brd4 vs Brd4-pm P = 0.0085, t = 2.488). (f) Pearson correlation coefficient for H4K16acetyl colocalization with Brd4 with CK2 site mutations (two-sided two-tailed t test for Brd4 n = 35 neurons, for Brd4 + BDNF n = 21, for S492A n = 29, and for Brd4-pm n = 18 from 3 biological replicates, for Brd4 vs Brd4 + BDNF P = 0.0164, t = 2.477, for Brd4 vs S492A P = 0.00167, t = 3.286, for Brd4 vs SSS492A P = 0.035, t = 2.166. *, p<0.05. ***, p<0.001. a.u. arbitrary units. Full-length blots are presented in Supplementary Figure 11. p’ase, phosphatase. Error bars represent standard error. Scale bar is 10 μM.