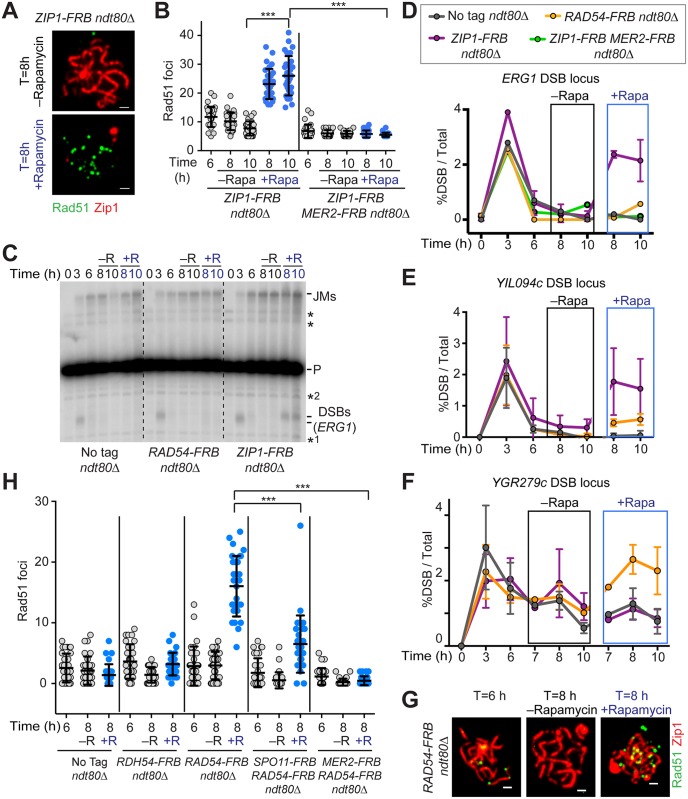
Fig 5. DSBs continue to form on synapsed chromosomes and require Rad54 for repair.
Rapamycin was added to part of the culture at T = 6 h for depletion of various FRB-tagged nuclear proteins. Samples were analyzed at the indicated time points in the presence or absence of the drug. (A) Rad51 (green) and Zip1 (red) immunofluorescence on spread chromosomes of ZIP1-FRB ndt80Δ (H7421) cells. Scale bar, 1 μm. (B) Quantification of the number of Rad51 foci per spread nucleus upon depletion of Zip1-FRB (H7421) or co-depletion of Zip1-FRB and Mer2-FRB (H8359). n = 30; error bars are S.D. from the mean; *** p < 0.001 Mann-Whitney test. (C) Southern analysis to monitor DSBs at the ERG1 locus in control cells (H7137) and before/after depletion of Rad54-FRB (H7121) or Zip1-FRB (H7421). P, parental unbroken fragment; JM, joint molecule repair intermediates; * nonspecific bands; *1 and *2 were used as anchors to measure the arbitrary molecular mass of DSB fragments in (S6C Fig). (D, E, F) Percentage of DSB fragments over total DNA at the ERG1 (D), YIL094c (E) and YGR279c hotspot locus (F) for the indicated genotypes, time points and treatments. (G) Rad51 (green) and Zip1 (red) immunofluorescence on spread chromosomes of RAD54-FRB ndt80Δ (H7121) cells. Scale bar, 1 μm. (H) Quantification of the number of Rad51 foci per spread nucleus in control strains (H7137) or upon depletion of Rdh54-FRB (H7485), Rad54-FRB (H7121) or co-depletion of Rad54-FRB with Spo11-FRB (H7740) or Mer2-FRB (H7840). n = 30; error bars are S.D. from the mean; *** p < 0.001 Mann-Whitney test.