Abstract
Histone phosphorylation has a profound impact on epigenetic regulation of gene expression, chromosome condensation and segregation, and maintenance of genome integrity. Histone H3 Serine 10 is evolutionally conserved and heavily phosphorylated during mitosis. To examine Histone H3 Serine 10 phosphorylation (H3S10ph) dynamics in mitosis, we applied immunogold labeling and confocal microscopy to visualize H3S10ph expression in MCF-7 cells. Confocal observations showed that MCF-7 cells had abundant H3S10ph expression in prophase and metaphase. In anaphase the H3S10ph expression was significantly decreased, and displayed only sparsely localized staining that mainly associated with the chromatid tips. We showed that immunogold bead density distribution followed the H3S10ph expression patterns observed in confocal analysis. At a higher magnification in metaphase, the immunogold beads were readily visible and the bead distribution along the condensed chromosomes was distinctive, indicating the specificity and reliability of the immunogold staining procedure. In anaphase, the beads were found to distribute focally in specific regions of chromatids, reinforcing the confocal observations of differential H3 phosphorylation. This is the first report to show specific H3S10ph expression with immunogold technique. Additionally, with confocal microscopy we analyzed H3S10ph expression in an immortalized cell line derived from benign uterine smooth muscle tumor cells. H3S10ph epitope was expressed more abundantly during anaphase in the benign tumor cells, and there was no dramatic differential expression within the condensed chromatid clusters as observed in MCF-7 cells. The differences in H3S10ph expression pattern and dynamics may contribute to the differential proliferative potential between benign tumor cells and MCF-7 cells.
INTRODUCTION
Eukaryotic cells must appropriately respond to a very diverse set of extracellular stimuli to maintain precise control of proliferation, differentiation, and apoptosis. These stimuli originate at the cell surface and are transduced via genomic signaling pathways to the nucleus or via nongenomic signaling pathways to targeted organelles. One of the major post-translational modifications required for various signaling pathways is protein phosphorylation. There is strong evidence to suggest that histone H3 phosphorylation serves as an essential component for diverse complex signaling pathways (Hans et al., 2001). Certain histone H3 modifications, such as phosphorylation at Serine 10 (Ser10), appear to have both positive and negative impact on transcriptional activation and chromosome condensation (Wei et al., 1999). In interphase, phosphorylation of histone H3 at Ser10 (H3S10ph) correlates with chromatin relaxation and transcription activation; whereas, in mitosis H3S10ph is correlated with chromatin condensation. The H3S10ph is mediated by several kinases, such as MSK1/2 (mitogen and stress-activated protein Kinases 1 and 2), cAMP-dependent protein kinase A (PKA), Aurora Kinase, and ribosomal S6 kinase 2 (RSK2) (Oki et al., 2007). The aberrant expressions of these kinases have been linked to multiple human diseases. Overexpression of Aurora B kinase is associated with colorectal cancer due to enhanced phosphorylation of histone H3 (Ota et al., 2002); whereas, a reduction of RSK2 levels interferes with neuronal precursor cell differentiation and may be the cause of the cognitive deficits observed in Coffin-Lowry Syndrome (Dugani et al., 2010). It has been shown that mutation of the histone H3 tail at Serine 10 impairs chromosome condensation and segregation in vitro (Wei et al., 1999).
Environmental stressors have direct impacts on posttranslational covalent modifications of histone tails including phosphorylation, methylation, acetylation, and ubiquitination. These types of modification dramatically alter chromatin structure and modulate DNA processes such as cell cycle, gene transcription, and silencing. In contrast to DNA methylation, the covalent modification of the histone tail is far more complex and dynamic (Reichard et al., 2012). The MAP kinase cascades are considered to be the key players in nucleosomal responses leading to H3S10ph and activation of immediate early (IE) response genes such as Fos and Jun in response to environmental agents (Thomson et al., 1999).
Several environmental factors such as nickel (Ke et al., 2008), cigarette smoke (Ibuki et al., 2014), arsenite (Dong et al., 2006), and ultraviolet light (Zhong et al., 2000) have direct effects on H3S10ph. Nickel phosphorylated H3S10 via the JNK pathway has been implicated in carcinogenesis (Ke et al., 2008). Cigarette smoke has the potential to enhance cell proliferation, survival, anchorage-independent growth, and tumor growth through its ability to promote H3S10ph in the promoter regions of oncogenes such as FOS and JUN, which is mediated mainly via JNK pathways and PI3K/ Akt pathway (Ibuki et al., 2014). The activation of Fos and Jun promoters by H3S10ph is reported to be critical for neoplastic cell transformation (Choi et al., 2005; Kim et al., 2008). Arsenic is a potent carcinogen and a strong inducer of H3 phosphorylation mediated through MAP kinase pathways, extracellular signal-regulated kinases (ERKs) and p38 kinase in interphase (He et al., 2003). It was shown that arsenite induced phosphorylation of histone H3 in a time- and dose-dependent manner (Dong et al., 2006). Evidently, environmental stressors including toxic chemical exposures have significant impacts on phosphorylation modification of the histone H3 tail that are associated with cell proliferation and in some instances cancer development. H3S10ph may serve as a potential indicator for evaluating the mitogenic and/or tumorigenic potential of environmental compounds.
Therefore, to assess the localization of the histone tail modification marker H3S10ph in chromatin structures, we developed reliable and quick experimental procedures to visualize its expression with high resolution. In this study, we have successfully implemented experimental protocols to visualize H3S10ph with immunogold labeling and transmission electron microscopy, and confocal imaging in immortalized benign and malignant human cell lines.
MATERIALS AND METHODS
Cells and Reagents
Human breast cancer MCF-7 cells were cultured in DMEM medium supplemented with 10% FBS. The human uterine leiomyoma cell line (ht-UtLM), immortalized via retroviral transfection (PLX1N vector, CLONTECH) with human telomerase, was previously generated in our laboratory (Carney et al., 2002). G418 Sulfate (Geneticin® Selective antibiotic, Life Technologies Cat# 10131-035) was added to ht-UtLM cell culture media at a dose of 1.0 μl/ml (Carney et al., 2002). Both cultures were kept in a standard tissue culture incubator at 37°C with 5% CO2.
Immunofluorescence and Confocal Microscopy
Immunofluorescence staining was performed to detect H3S10ph. Both MCF-7 and ht-UtLM cells were grown in glass bottom microwell dishes (MatTek Corporation, Part No# P35G-1.5-14-C). Approximately 50,000 cells were introduced into each dish, and grown for 2-3 days at 37oC in a CO2 incubator to reach 80% confluence. The cells were fixed in methanol for 5 min on ice, blocked with 10% normal goat serum for 20 min on ice, and incubated with Phospho-Histone H3 (Ser10) antibody (Cell Signaling Cat# 9701S) diluted 1: 250 with 1.5% normal goat serum in PBS at 4oC overnight. The cells were then incubated with Alexa Fluor® 488 goat anti-rabbit IgG (H+L) antibody (1:3000 dilution, Life Technologies Cat# A11008) at room temperature for 1 hr, and counterstained with 100 ng/ml 4,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Molecular Probes Cat# D1306) for 30 min. Confocal images were taken on a Zeiss LSM710-UV Confocal Microscope (Carl Zeiss) using a Plan-Apochromat 63X/1.40 oil DIC M27 objective.
Immunogold Labeling and Transmission Electron Microscopy
For transmission electron microscopy and immunogold labeling studies, cells were maintained in culture until 90% confluent. The cells were detached with 0.25% trypsin solution (Life Technologies Cat# 25200-056), resuspended in 1X PBS and centrifuged at 1,000 rpm for 2 min. The supernatant was removed and the cell pellet resuspended in 5 ml 4% Paraformaldehyde in 0.1 M Phosphate buffer, pH 7.2, overnight. The next day, the fixed cell pellets were brought to the Electron Microscopy lab for further processing. Initially, the cell pellets were embedded in 4% molten agar (temperature maintained at approximately 50°C) and the agar-containing cell pellet was allowed to solidify at room temperature. The agar-embedded block was trimmed and prepared for processing in a Leica® EM TP automatic tissue processor. Briefly, the block was rinsed in 0.1 M Phosphate Buffer (pH 7.2), dehydrated through a series of graded alcohols, and infiltrated and embedded in a gelatin capsule (Electron Microscopy Sciences, Cat# 70100) with LR White Embedding Resin (Electron Microscope Sciences, Cat# 14381-CA). The LR white infiltrated cell pellet was placed in a 55°C to 60°C oven overnight for polymerization. The polymerized blocks were trimmed and thick-sectioned (500 - 700 nm thick), and stained with Toluidine blue. The thick sections were examined by light microscopy for selection of the blocks to be thin-sectioned (70 - 90 nm thick). The thin sections were subsequently collected on 200 mesh formvar-coated Nickel grids (Electron Microscope Sciences, Cat# FF200-Ni-50).
For the immunogold labeling, the nickel grids were first washed in 1x PBS for 10 min, and blocked with blocking buffer (10% normal goat serum and 1% FBS in 1x PBS) for 30 min. The nickel grids were incubated overnight at 4°C with the primary antibody, Phospho-Histone H3 Ser10 (same as used for confocal experiments), at a 1:50 dilution or normal rabbit IgG at a comparable dilution to the primary antibody (served as negative control) in 1x PBS containing 10% normal goat serum. After incubation, the grids were washed 10 times with PBS buffer in a sterile petri dish. The grids were then incubated with goat anti-rabbit IgG H&L coupled to 10-nm colloidal gold particles (Abcam Inc. Cat# ab27234) at a dilution of 1:80 with 10% normal goat serum in 1x PBS for 2 h at room temperature. The nickel grids were then washed 10 times with 1x PBS and dried on filter paper overnight at room temperature. To reduce the nonspecific immunogold precipitation in the cells, all the buffers used in the immunogold labeling procedure were filtered with a 0.22 μm filter unit in advance. The grids were observed with an electron transmission microscope (Tecnai G2 12 Biotwin microscope, FEI™).
Cell Proliferation Rates and Proliferating Cell Nuclear Antigen (PCNA) Labeling Indices
The cell proliferation rates were quantified by counting the number of cell nuclei in a given area at various time points. Fifty-thousand cells for each cell line were pipetted into glass-bottom petri dishes and incubated at 37°C, and allowed to attach overnight. At 0, 24, 48, and 72 h, the cells were fixed with cold methanol for 5 min and stained with DAPI. Cell culture confocal images were taken as described above. For each time point, three images were taken. The nuclear counts for each image, corresponded to the size of 1.766 mm2 of the glass-bottom dish, which was done with MetaMorph Microscopy Automation and Image Analysis Software (v.7.8.60, ©1992-2014, Molecular Devices, LLC). Data are shown in Supplemental Figure 2. For PCNA labeling, 25,000 ht-UtLM or MCF-7 cells / slide were added to single-well (10 cm2) chamber slides (Thermo Scientific #177453). Serum free DMEM-F12 without phenol red replaced the culture medium for 24 h. The cells were placed in regular DMEM-F12 cell culture medium with 10% serum for 24, 48, and 72 h. Cells were fixed with 70% of ethanol and 4% of paraformaldehyde for 10 min. Cell permeation was done with 0.2% Triton X-100 (Sigma St. Louis MO, Cat# T-9284) and a peroxide block (BioGenex, Fremont California Cat# HK111-5K) was used for quenching endogenous peroxidase activity. Cells were incubated with primary anti-mouse (PCNA) antibody (Cell Signaling Technology, Cat#2586) diluted at 1:2000 for 2 hr. The negative control was a normal mouse serum (Jackson Immunoresearch Laboratories, Inc.). A goat anti-mouse, IgG chain, biotinylated secondary antibody (Jackson Immunoresearch Laboratories, Inc. HSVMS Cat# BA-2001) was applied at a 1:800 dilution for 30 min. A label was used on the slides for 30 min (Super Sensitive HPR label, BioGenex, #HK330-9K) and a chromogen, DAB (BioGenex, #HK124-5K) for 10 min. The counterstain was done for 20 s using Harris’ Hematoxylin. Ethanol at concentrations of 95% and 100% was used to dehydrate the slides. Percent PCNA labeling was determined by the number of cells having positively stained (brown) nuclei divided by 1,000 cells total (i.e., labeled and unlabeled) and multiplied by 100 (Supplemental Fig. 2). The data for nuclear cell counts and PCNA labeling were statistically analyzed using an analysis of variance followed by a Dunnett's test to compare each time point with time 0 within each cell line. Two-sample t-tests were used to compare between cell lines at each time point. All P-values are two-sided.
RESULTS
Detection of H3S10ph Epitope Expression During Mitosis in MCF-7 Cells by Immunofluorescence and Confocal Microscopy
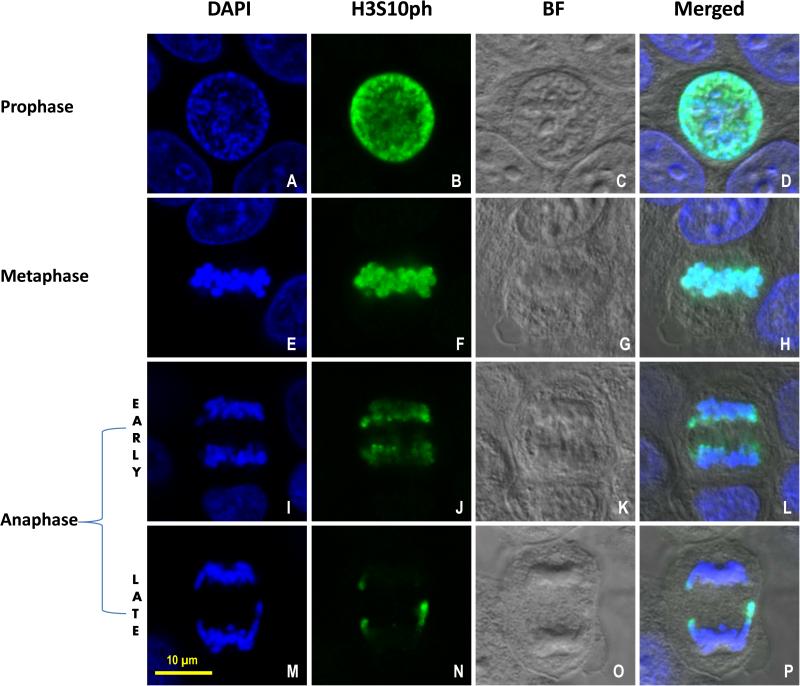
As shown in Figures 1A-D, the H3S10ph antibody stained MCF-7 cells intensely and specifically in prophase within the nucleus. In MCF-7 cells at prophase, the nuclei usually were round with an average diameter of approximately 15 μm, contained a distinctive nuclear membrane, and occupied the majority of the cellular volume. At the beginning of metaphase, the nuclear envelope is disrupted and broken into small membrane vesicles. The images in Figure 1E-H depict an MCF-7 cell in metaphase with chromosomes congressed at the metaphase plate or spindle equator. At this stage, no clear nuclear membrane is visible. The H3S10ph signal was highly expressed (Fig. 1F) and mostly colocalized with DAPI in the congressed chromosomes (Fig. 1H). At early anaphase, as demonstrated in the MCF-7 cell shown in Figure 1I-L, the two daughter-chromatid clusters began to separate. The intensity of the H3S10ph signal in early anaphase was reduced compared to that of metaphase (Fig. 1J). The majority of the daughter-chromatid cluster was blue, indicating lack of H3S10ph expression or decreased H3S10ph epitope expression in the blue regions (Fig. 1L). Interestingly, we observed a moderate amount of H3S10ph signal existed almost universally on the front edge of the daughter chromatid mass facing the spindle equator in early anaphase. During late anaphase, as shown in Figure 1M-P, the two chromatid clusters were further separated. The H3S10ph signal was significantly reduced or disappeared at the main body of the daughter chromatid mass (Fig. 1N). Notably, the H3S10ph signal at the front edge of the separated chromosome mass was also almost completely diminished. The only signals detectable at this stage were located at the lateral tips of the daughter chromatids. The three green dots representing H3S10ph epitope expression (Fig. 1N) in the green channel at the late anaphase stage were easier to discern than when merged with the DAPI channel (Fig. 1P).
Figure 1.
H3S10ph expression during mitosis in MCF-7 cells. Confocal images of MCF-7 cells representing prophase (A, B, C, D), metaphase (E, F, G, H), early anaphase (I, J, K, L) and late anaphase (M, N, O, P) during mitosis. Blue signal in the first column (A, E, I, M) = DAPI; Green signal in the second column (B, F, J, N) = Histone H3 phosphorylation at Serine 10 (H3S10ph); the third column (C, G, K, O) = Bright field (BF); the fourth column (D, H, L, P) = merged image. Scale bar = 10 μm.
Confirmation of Specificity of H3S10ph Labeling by Immunogold Electron Microscopy in MCF-7 Cells
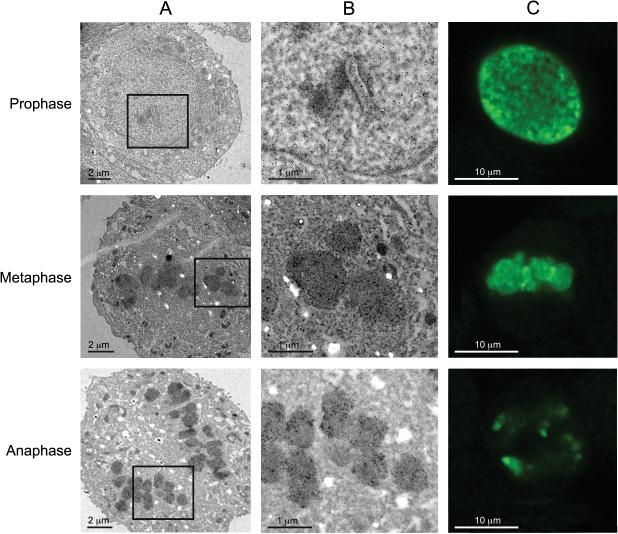
Immunogold labeling and transmission electron microscopy has certain advantages over conventional confocal assays, as the localization and distribution of the antigen to specific cellular organelles and structures can be clearly observed at the ultrastructural level with high resolution. The immunogold labeling procedure was performed to evaluate the density and specific localization of H3S10ph signal in MCF-7 cells at a high resolution. As demonstrated in Figure 2A, at a low magnification, the majority of the cell is included in the figure and the overall chromosome or chromatid organizational patterns are shown in the transmission electron micrographs of cells in each phase. At prophase, immunogold beads were detected in association with fibrils in the nuclear matrix and at the nuclear periphery of cells having an intact and discernable nucleus.. The condensed chromosomes at metaphase and anaphase were labeled with distinctive gold beads with higher density than in prophase. In metaphase, the gold bead distribution followed the congressed chromosomes along the equator plane, and in anaphase the gold bead distribution followed the two chromatid clusters in the process of being pulled apart to the respective centrosomes. In the lower magnification electron micrographs, the individual chromosomes or individual chromatids were readily distinct, which could not be detected in the confocal images. At a higher magnification (Fig. 2B), individual immunogold beads could be observed in all of the three phases. At metaphase, the immunogold beads were somewhat diffusely distributed over the congressed chromosome sections, and there were minimal focal regions of densely aggregated beads (Figs. 2B and 3A). At anaphase, the overall density of the immunogold beads was reduced in most of the chromatids compared to metaphase. We observed that some cells showed varying immunogold labeling densities within individual chromatids at anaphase (Fig. 2B). In a separate case, we observed that at anaphase one chromatid was intensely labeled with gold beads, resulting in a localized electron-dense region where the gold bead density was estimated to be at least 5 times more than those of the adjacent chromatids (Fig. 3B). This type of high expression of the H3S10ph signal in a specific chromatid at late anaphase was also observed in the confocal studies in the MCF-7 cells (see Figs. 1N, P and 2C).
Figure 2.
H3S10ph immunogold staining showing MCF-7 cells in prophase, metaphase and anaphase. (A) At low magnification, dark staining was seen in prophase, metaphase and anaphase, which may represent the immunogold bead signals. (B) At high magnification, immunogold beads can be resolved in prophase, metaphase and anaphase. Note the immunogold bead density gradient within a chromatid cross-section in anaphase. (C) Confocal Green channel displaying H3S10ph signal at prophase, metaphase and anaphase. A, Scale bar = 2 μm; B, Scale bar = 1 μm; C, Scale bar = 10 μm.
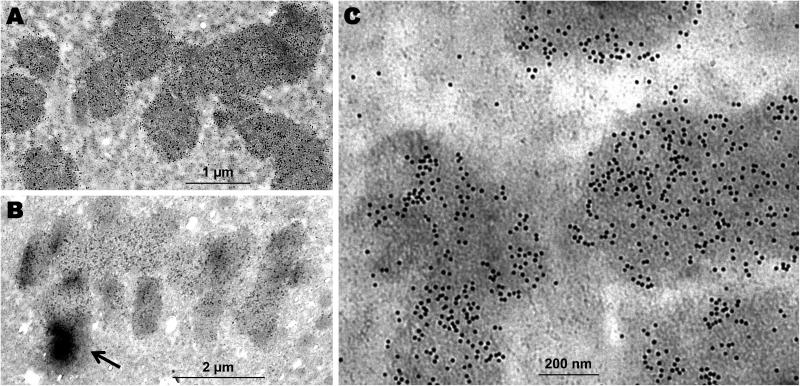
Figure 3.
MCF-7 cells immunogold stained with H3S10ph antibody. (A) The 10 nm immunogold beads distributed relatively evenly along the condensed chromosomes at metaphase. (B) The immunogold bead density distribution displayed dramatic gradients (arrow) among chromatids at anaphase. (C) Immunoelectron microscopy image to show the portion of chromosomes at metaphase with distinctive 10 nm gold beads. A, Scale bar = 1 μm; B, Scale bar = 2 μm; C, Scale bar = 200 nm.
At an even higher magnification, as shown in Figure 3C, in metaphase the H3Ser10ph epitope signal appeared as discrete and distinctive spherical beads that were distributed relatively evenly along the condensed chromosomes. This was consistent with the confocal observation for MCF-7 cells at metaphase, in which the H3S10ph signal distribution along the congressed chromosomes showed reduced varying density gradients in comparison to anaphase (Fig. 1F and 1H). Gold beads were also detected in the cytoplasm adjacent to regions of condensed chromosomes, although the gold bead density in these regions was considerably less than in the condensed chromosomes and most likely due to loss of nuclear membrane integrity (Fig. 3C).
Comparison of the Expression of H3S10ph During Mitosis in a Benign Human Cell Line by Immunofluorescence Confocal Microscopy
The H3S10ph expression pattern in benign uterine leiomyoma cells (ht-UtLM cells) during mitosis is demonstrated in Supplemental Figure 1. There were several major differences in H3S10ph expression dynamics between the two cell lines. At prophase, the nuclei of ht-UtLM cells were classically, cigar-shaped (Supplemental Fig. 1A-D). At this stage, H3S10ph was expressed moderately in the nucleus. At metaphase, the chromosomes converged at the plate halfway between the poles, and H3S10ph epitope was highly expressed and relatively uniformly present in the packed chromosome mass (Supplemental Fig. 1E-H).
At early anaphase shown in Supplemental Figure 1I-L, the chromatids were pulled apart toward the poles to form two chromatid clusters on each side of the equator. The H3S10ph epitope was highly expressed; however, there was differential expression in the chromatid clusters with the edge displaying the highest H3S10ph expression (Supplemental Fig. 1J, and L). It was a consistent observation that in the merged channel image, the center of the chromosome cluster was blue, hinting that there was no or low H3S10ph expression (Supplemental Fig. 1L). At late anaphase, the two chromosome clusters were pulled apart further from the equator without formation of a distinct nuclear membrane in the daughter cells (Supplemental Fig. 1M-P). In the ht-UtLM cells cytokinesis occurred at this stage. During cytokinesis, the cell membrane approximately located in the middle of the cell, perpendicular to the spindle axis and adjacent to the metaphase plate, was drawn inward to form a cleavage furrow. At late anaphase, in ht-UtLM cells, both the chromatid clusters still had moderately intense H3S10ph epitope expression, mostly around the edges of the clusters (Supplemental Fig. 1N and P). Similar to the observation in early anaphase, the center of the chromosome clusters remained blue, indicating the low or lack of H3S10ph epitope expression.
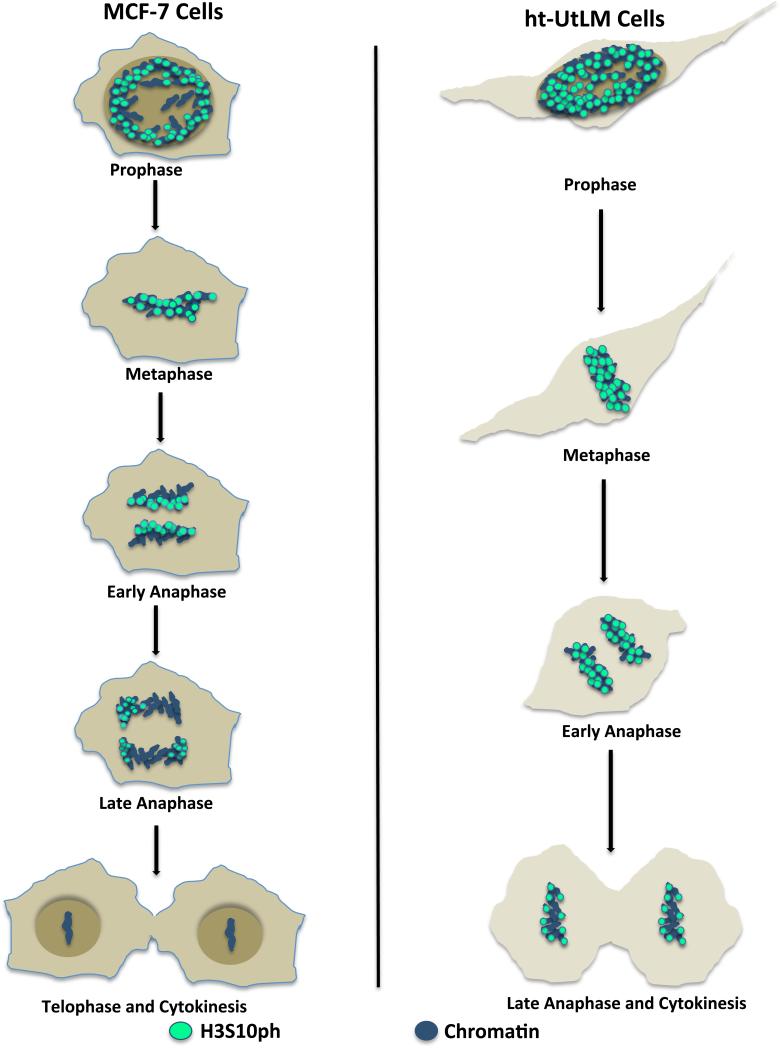
When the H3S10ph expression patterns in different mitotic stages were compared between MCF-7 and ht-UtLM cells, it was not difficult to discern the differences as illustrated in Figure 4. Notably, in early and late anaphase, MCF-7 cells had significantly reduced H3S10ph expression in chromatid clusters, possibly suggesting more potent dephosphorylating activity than ht-UtLM cells (Fig. 1J, L, N and P; Supplemental Fig. 1J, L, N and P). It appeared that in MCF-7 cells, reduction in H3S10ph epitope expression started at the edge of the chromatid cluster closest to the spindle pole and moved toward the edge adjacent the spindle equator. In contrast, reduction of H3S10ph signal in ht-UtLM cells started in the center of the chromatid clusters. In late anaphase, ht-UtLM cells still had moderate expression of the H3S10ph epitope in the outer boundaries of the chromatid clusters, whereas MCF-7 cells only displayed much-reduced H3S10ph signal that only occurred focally at the tips of a few chromatids. When growth rates were compared between the two cell lines, MCF-7 cells showed significantly increased growth rates at 0, 24, 48, and 72 h and similarly proliferating cell nuclear antigen labeling indices were significantly higher in MCF-7 compared to ht-UtLM cells at all time points (Supplemental Fig. 2). Therefore, the temporal and spatial expression pattern of H3S10ph in mitosis appears to differ between MCF-7 and ht-UtLM cells, and these differences might account for the varied proliferation potential of the two cell lines.
Figure 4.
Comparison of H3S10ph expression dynamic during mitosis in MCF-7 and ht-UtLM cells. The two cell lines have different spatial and temporal distribution of H3S10ph during mitosis.
DISCUSSION
Histone H3 tail at Serine 10 is globally phosphorylated and dephosphorylated in all eukaryotes in mitosis and meiosis, but the function of this specific post-translational modification in cell cycle control has been the subject of debate. Generally, it is believed that phosphorylation of the histone H3 tail is a part of a complex signaling mechanism. In late G2 phase, phosphorylation of histone H3 starts to occur on pericentromeric heterochromatin. As mitosis proceeds, the phosphorylation spreads along the chromosome. The condensed chromosomes are phosphorylated almost completely at prophase and maintain through metaphase. Dephosphorylation of histone H3 begins in anaphase and ends at early telophase. A strong correlation between the initial chromatin condensation and H3 tail phosphorylation has been established (Hans et al., 2001). Phosphorylation modifications on histones can contribute significant negative charges to the histone that can influence chromatin structure and transcriptional activity. The extent of phosphorylation on histones is regulated by kinases and phosphatases that add and remove phosphate groups (Sugiyama et al, 2002).
In this study, we observed several unique temporal and spatial features for H3S10ph expression in MCF-7 cells. At metaphase, the congressed chromosome cluster at the equator plane displayed strong H3S10ph signal; however, at this stage the H3S10ph epitope distribution over the chromosome mass was not homologous. At metaphase, the MCF-7 cells had green, aqua and blue patches present on congressed chromosomes, suggesting that the condensed chromosomes were differentially phosphorylated at the H3 tail. At early anaphase, the H3S10ph epitope signal was globally reduced in the MCF-7 cells, but a moderate amount of H3S10ph signal existed generally along the front edge of the chromatid mass facing the spindle equator. In late anaphase, H3S10ph signal was reduced further on the pulled chromatid clusters; however, strong H3S10ph expression was still detected at the tips of selected chromatids, suggesting the presence of a dramatic H3S10ph epitope signal gradient on the chromatid clusters. The heterologous H3S10ph epitope signal distribution on condensed chromosomes in MCF-7 cells was also independently observed with immunogold labeling and transmission electron microscopy evaluations. These observations suggested that the kinase and phosphatase responsible for regulating H3S10ph expression and dephosphorylation on the condensed chromatids might have differential distribution over the condensed chromosomes. Interestingly, in ht-UtLM cells that are derived from a human uterine leiomyoma, the H3S10ph expression pattern at metaphase and anaphase was quite different. The H3S10ph epitope showed relatively uniform expression on the congressed chromosome cluster at metaphase, without noticeable blue patches found on the cluster. At anaphase, the dephosphorylation effect was first noticed in the center of the pulled chromatid clusters and the remaining H3S10ph signals occurred mostly surrounding the chromatid cluster bodies. It is reasonable to assume that the kinase / phosphatase enzyme system responsible for regulating H3S10ph epitope expression on the condensed chromosomes are distributed differentially between MCF-7 and ht-UtLM cells.
Chromosomal segregation during mitosis is regulated by kinases and phosphatases, notably the so-called Aurora kinase and protein phosphatase 1 axis. Aurora B kinase, being a chromosomal passenger protein, is responsible for H3S10ph during mitosis (Meadows, 2013). It is known that Aurora B kinase, which is responsible for H3S10ph, translocates to the central mitotic spindle and is highly enriched there in anaphase. In comparison to ht-UtLM cells, the chromatid clusters in MCF-7 cells would be closer to the region where Aurora B kinase is proposed to be highly enriched. However, MCF-7 chromatid clusters at both early and late anaphase lost their H3S10ph signal more extensively. Even though the chromatid clusters in MCF-7 cells at anaphase were more closely positioned to the central mitotic spindle, which is reportedly a rich source of Aurora B kinase, the phosphorylation status could not be well maintained. Therefore, alternative factors such as unique PP1 distribution pattern along the chromosomes in anaphase, may account for the relatively faster disappearance of H3S10ph signal in MCF-7 cells compared to ht-UtLM cells during anaphase. This hypothesis needs to be confirmed by further investigations of Aurora B kinase and PP1 expression dynamics during mitosis in these two cell lines.
In this study, when growth rates were compared between the two cell lines, MCF-7 cells showed significantly increased growth at all time points evaluated, and similarly proliferating cell nuclear antigen labeling indices were significantly higher in MCF-7 compared to ht-UtLM cells. Also, the temporal and spatial expression pattern of H3S10ph in mitosis differed between MCF-7 and ht-UtLM cells, and we hypothesize these differences might account for the varied proliferation potential of the two cell lines. Recently, the utility of H3S10ph as an indicator of mitosis in breast cancer grading has been explored by Cui et al (2015). These investigators have found that H3S10ph staining of breast biopsy samples demonstrates discrete, strong nuclear immunoreactivity in mitotically active cells, with numerous advantages over conventional hematoxylin and eosin (H&E) and Ki-67 staining procedures. The advantages have prompted the reconsideration of breast cancer grading with the integration of H3S10ph staining. The effective detection of mitotic figures with H3S10ph staining has also been seen with cutaneous melanoma and has been projected to have profound implications for cancer staging and patient management of (Tetzlaff et al, 2013).
Modulation of histone H3 by phosphorylation events may also serve as a promising area to explore when evaluating epigenetic mechanisms of environmental agents. There is emerging interest in how environmental agents and external stimuli can modulate the kinase and phosphatase activities of histones and regulate phosphorylation, based on the observation that kinases receiving signals from external cues can enter the nucleus and modify histones. The direct signaling from external or environmental stimuli to chromatin and epigenetic modification of histones by phosphorylation events may warrant the rationale for studying chromatin-modifying enzymes as environmental targets. Our immunogold labeling and confocal microscopy analyses have offered high-resolution details pertaining to the expression dynamics of the H3S10ph epitope in these two cell lines during mitosis. Our studies have demonstrated that the H3S10ph signal is differentially expressed in MCF-7 condensed chromosomes, and significant differences in H3S10ph dynamics exist between a benign mesenchymal tumor cell and malignant breast epithelial cell. This study has provided necessary background information defining H3S10ph expression dynamics during the cell cycle in MCF-7 and ht-UtLM cells. The data derived from these studies warrant further evaluation of the impact of endocrine disrupting and other environmental agents, such as bisphenol A (BPA), BPS, Tetrabromobisphenol A (TBBPA), cigarette smoke condensate, cadmium and arsenic, on H3 phosphorylation and mitosis. It is reasonable to postulate that the unique pattern of H3S10ph expression we observed during mitosis may regulate the differential cellular proliferative potential of benign versus malignant cells, and that epigenetic regulation of genes by histone modification through phosphorylation marks may be a target of environmental toxicants.
Supplementary Material
ACKNOWLEDGMENTS
The authors kindly thank Drs. Gordon Flake and Ronald Herbert for their critical review of this manuscript. The authors would like to also thank Mr. Charles J. Tucker and Dr. Agnes K. Janoshazi from NIEHS for their excellent guidance with confocal microscopy, and Dr. Grace E. Kissling, NIEHS, NTP for her assistance with statistical analysis of data. The authors greatly appreciate the technical assistance of Mrs. Beth Mahler and Mrs. Lois Wyrick in compilation of the electron microscopy image plates.
Supported by: The Intramural Research Program of the NIH, NIEHS and DNTP.
REFERENCES
- Carney SA, Tahara H, Swartz CD, Risinger JI, He H, Moore AB, Haseman JK, Barrett JC, Dixon D. Immortalization of human uterine leiomyoma and myometrial cell lines after induction of telomerase activity: molecular and phenotypic characteristics. Lab Invest. 2002;82:719–28. doi: 10.1097/01.lab.0000017499.51216.3e. [DOI] [PubMed] [Google Scholar]
- Choi HS, Choi BY, Cho YY, Mizuno H, Kang BS, Bode AM, Dong Z. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 2005;65:5818–5827. doi: 10.1158/0008-5472.CAN-05-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Harada S, Shen D, Siegal GP, Wei S. The Utility of Phosphohistone H3 in Breast Cancer Grading. Appl Immunohistochem Mol Morphol. 2015;23:689–695. doi: 10.1097/PAI.0000000000000137. [DOI] [PubMed] [Google Scholar]
- Dong Z, Bode AM. The role of histone H3 phosphorylation (Ser10 and Ser28) in cell growth and cell transformation. Mol Carcinog. 2006;45:416–421. doi: 10.1002/mc.20220. [DOI] [PubMed] [Google Scholar]
- Dugani CB, Paquin A, Kaplan DR, Miller FD. Coffin-Lowry syndrome: a role for RSK2 in mammalian neurogenesis. Dev Biol. 2010;347:348–359. doi: 10.1016/j.ydbio.2010.08.035. [DOI] [PubMed] [Google Scholar]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- He Z, Ma WY, Liu G, Zhang Y, Bode AM, Dong Z. Arsenite-induced phosphorylation of histone H3 at serine 10 is mediated by Akt1, extracellular signal-regulated kinase 2, and p90 ribosomal S6 kinase 2 but not mitogen- and stress- activated protein kinase 1. J Biol Chem. 2003;278:10588–10593. doi: 10.1074/jbc.M208581200. [DOI] [PubMed] [Google Scholar]
- Ibuki Y, Toyooka T, Zhao X, Yoshida I. Cigarette sidestream smoke induces histone H3 phosphorylation via JNK and PI3K/Akt pathways, leading to the expression of protooncogenes. Carcinogenesis. 2014;35:1228–1237. doi: 10.1093/carcin/bgt492. [DOI] [PubMed] [Google Scholar]
- Ke Q, Li Q, Ellen TP, Sun H, Costa M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis. 2008;29:1276–1281. doi: 10.1093/carcin/bgn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Lee KW, Cho YY, Kang NJ, Oh SM, Bode AM, Dong Z. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 2008;68:2538–2547. doi: 10.1158/0008-5472.CAN-07-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JC. Interplay between mitotic kinesins and the Aurora kinase-PP1 (protein phosphatase 1) axis. Biochem Soc Trans. 2013;41:1761–1765. doi: 10.1042/BST20130191. [DOI] [PubMed] [Google Scholar]
- Ochi T, Nakajima F, Nasui M. Distribution of gamma-tubulin in multipolar spindles and multinucleated cells induced by dimethylarsinic acid, a methylated derivative of inorganic arsenics, in Chinese hamster V79 cells. Toxicology. 1999;136:79–88. doi: 10.1016/s0300-483x(99)00061-x. [DOI] [PubMed] [Google Scholar]
- Oki M, Aihara H, Ito T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell Biochem. 2007;41:319–336. [PubMed] [Google Scholar]
- Ota T, Suto S, Katayama H, Han ZB, Suzuki F, Maeda M, Tanino M, Terada Y, Tatsuka M. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002;62:5168–5177. [PubMed] [Google Scholar]
- Reichard JF, Puga A. Epigenetic histone changes in the toxicologic mode of action of arsenic. In: Sahu Saura C., editor. Toxicology and Epigenetics. Wiley; West Sussex: 2012. pp. 339–356. [Google Scholar]
- Sugiyama K, Sugiura K, Hara T, Sugimoto K, Shima H, Honda K, Furukawa K, Yamashita S, Urano T. Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene. 2002;21:3103–3111. doi: 10.1038/sj.onc.1205432. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyazaki K, Kita K, Ochi T. Trivalent dimethylarsenic compound induces histone H3 phosphorylation and abnormal localization of Aurora B kinase in HepG2 cells. Toxicol Appl Pharmacol. 2009;241:275–282. doi: 10.1016/j.taap.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Tetzlaff MT, Curry JL, Ivan D, Wang WL, Torres-Cabala CA, Bassett RL, Valencia KM, McLemore MS, Ross MI, Prieto VG. Immunodetection of phosphohistone H3 as a surrogate of mitotic figure count and clinical outcome in cutaneous melanoma. Mod Pathol. 2013;26:1153–60. doi: 10.1038/modpathol.2013.59. [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Zhong SP, Ma WY, Dong Z. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J Biol Chem. 2000;275:20980–20984. doi: 10.1074/jbc.M909934199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.