Extended Data Figure 1.
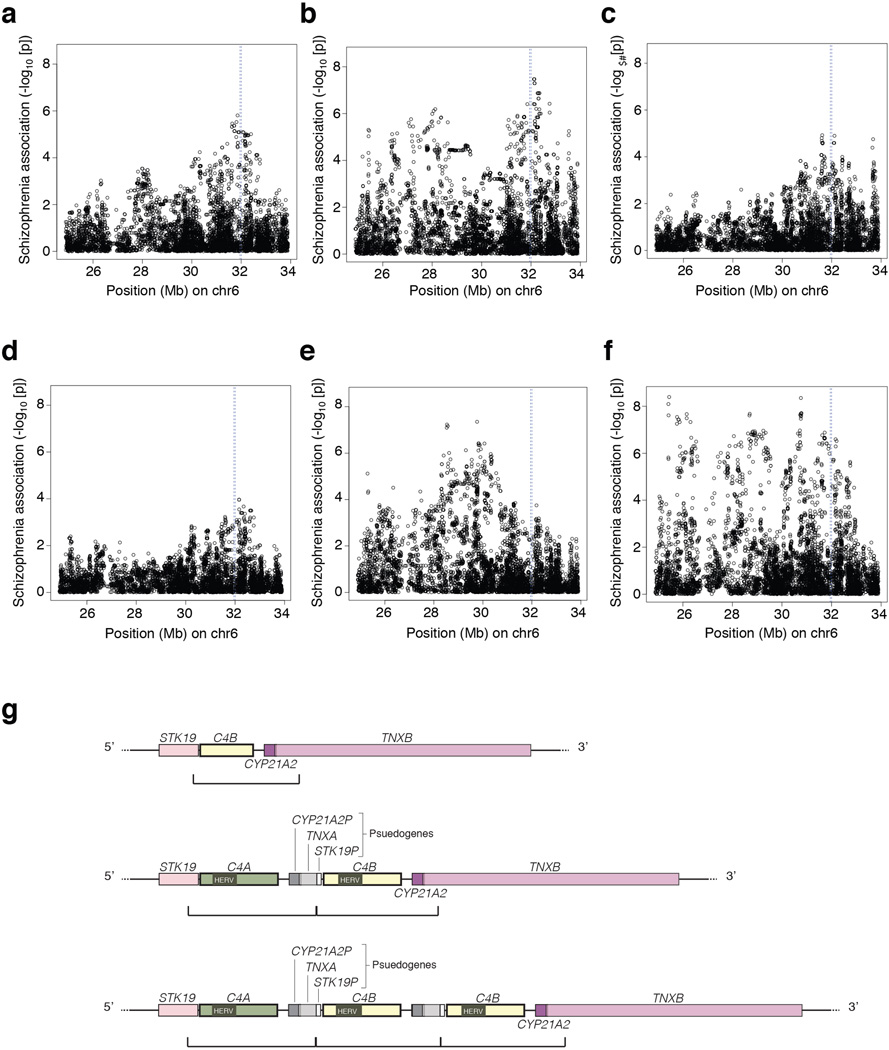
Association of schizophrenia to common variants in the MHC locus in individual case-control cohorts, and schematic of the repeat module containing C4.
(a–f) Data for several schizophrenia case-control cohorts that were genome-scanned before we began this work (a–d) exhibits peaks of association near chr6:32Mb (blue vertical line) on the human genome reference sequence (GRCh37/hg19). Note that association patterns vary from cohort to cohort, reflecting statistical sampling fluctuations and potentially fluctuations in allele frequencies of the (unknown) causal variants in different cohorts. Cohorts such as in (b), (e) and (f) suggest the existence of effects at multiple loci within the MHC region. Even in the cohorts with simpler peaks (a, c, d), the pattern of association across the individual SNPs at chr6:32 Mb does not correspond to the linkage disequilibrium (LD) around any known variant. This motivated the focus in the current work on cryptic genetic influences in this region that could cause unconventional association signals that do not resemble the LD patterns of individual variants.
(g) A complex form of genome structural variation resides near chr6:32 Mb. Shown here are three of the known alternative structural forms of this genomic region. The most prominent feature of this structural variation is the tandem duplication of a genomic segment that contains a C4 gene, 3’ fragments of the STK19 and TNXB genes, and a pseudogenized copy of the CYP21A2 gene. (This cassette is present in 1–3 copies on the three alleles depicted above; the boundaries below each haplotype demarcate the sequence that is duplicated.) Haplotypes with multiple copies of this module (middle and bottom) contain multiple functional copies of C4, whereas the additional gene fragments or copies denoted STK19P, CYP21A2P, and TNXA are typically pseudogenized. (Rare haplotypes with a gain or loss of intact CYP21A2 have also been observed18.) Note that although C4A and C4B contain multiple sequence variants, they are defined based on the differences encoded by exon 26, which determine the relative affinities of C4A and C4B for distinct molecular targets19,20 (Fig. 1). Many additional forms of this locus appear to have arisen by non-allelic homologous recombination and gene conversion (ref18 and Fig. 1).