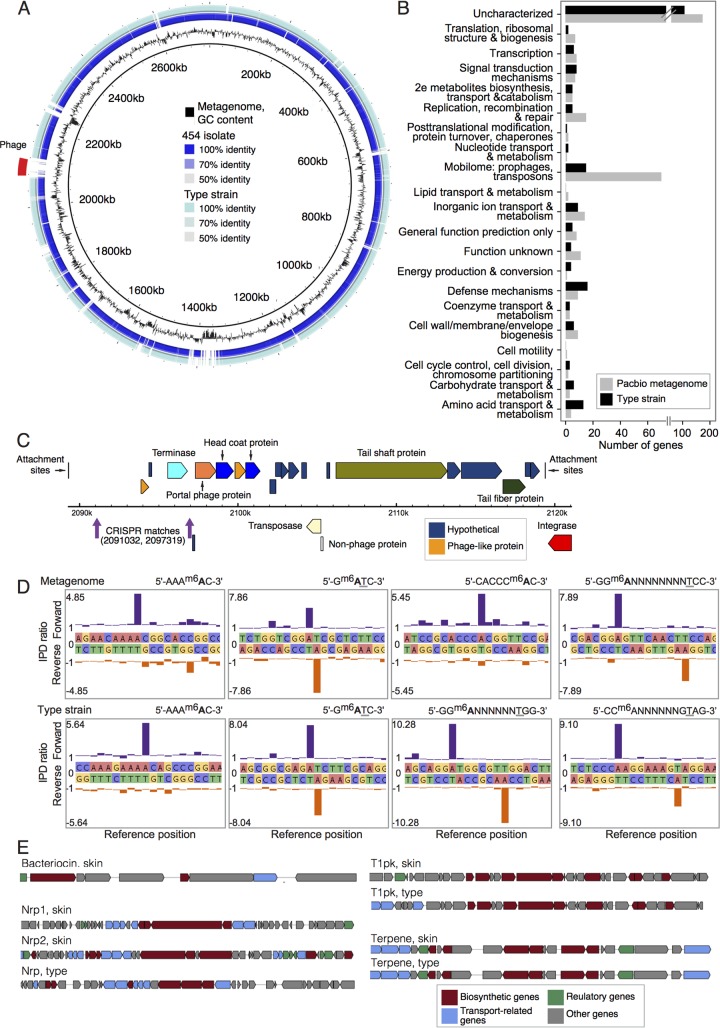
FIG 6 .
Select functional characterizations of C. simulans. (A) Whole-genome comparisons of the C. simulans genomes described in this study. The de novo C. simulans metagenome is used as a reference (innermost ring in black). The GC content of the metagenome is shown in the second ring in black. Ordered contigs from the 454-sequenced skin isolate are shown in the third ring. Alignment of the C. simulans type strain is shown in the fourth or outermost ring. Intensity of color shows the percent identity of the match. (B) COG categories of variable genes, those that are absent in either one of the two complete genomes. (C) Genome structure of the bacteriophage identified in panel A. The metagenome-derived C. simulans contains two CRISPR spacers that are a 100% match to this phage genome and are indicated in purple. (D) Epigenome analysis of C. simulans. Examples of kinetic modification detection signals of 6-methyladenine (m6A) in the C. simulans metagenome (top) and type strain (bottom). The x axis shows the template position and base calls, and the y axis shows the ratio of average interpulse durations (IPDs) for each DNA strand to the control. High deviations from the baseline level indicate a base modification, with the forward strand shown in purple and the reverse strand shown in orange. The methylated bases are indicated in bold type, and the underlined bases indicate methylation on the opposite DNA strand. (E) Examples of biosynthetic pathways predicted from the two complete genomes using antiSMASH 3.0.