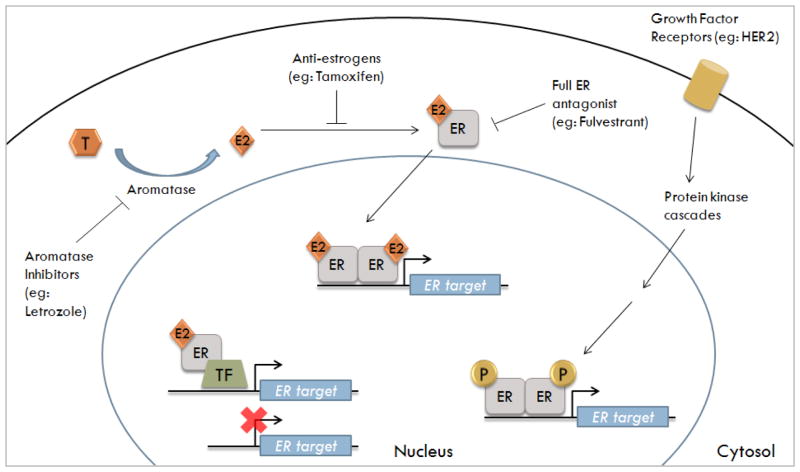
Figure 1. Three mechanisms of ER genomic signaling and the inhibition by antiestrogens and aromatase inhibitors.
Testosterone (T) is converted into estrogen (E2) by the enzyme aromatase. Normal breast cells synthesize E2 which has autocrine and paracrine functions. Breast cancer cells express higher levels of aromatase; thus, their E2 concentration is higher than normal breast cell. Furthermore, ER-positive breast cells require E2 for growth and utilize certain genomic signaling pathways to transcribe ER-regulated genes. These pathways include: classical genomic (E2-ER complex binds to the ERE); ERE-independent genomic (E2-ER complex binds to transcription factor-TF-binding sites); and non-classical genomic (ER is phosphorylated in absence of E2 via kinase cascades).