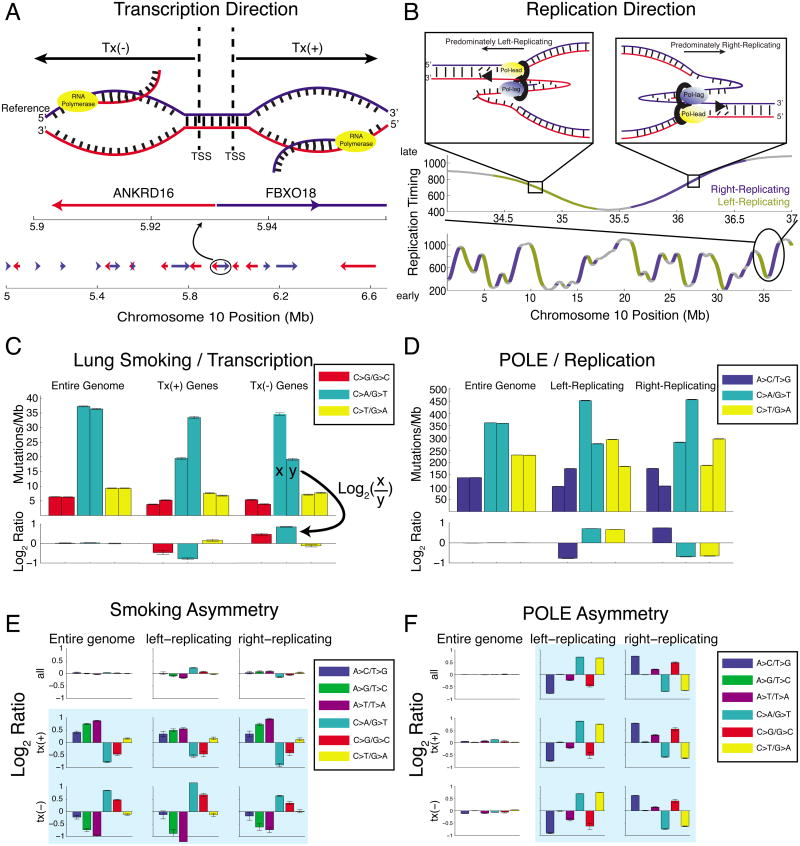
Figure 1. Mutational strand asymmetry associated with transcription (left) and replication (right).
(A) Transcription direction: Tx(+) regions carry the coding sequence of a gene on the genomic reference strand, and Tx(-) regions on the genomic complement strand. (B) Replication direction: positive slope in replication-timing data indicates general rightward movement of the replication complex (“right-replicating”), while negative slope indicates left-replicating. (C) Lung cancers show strong transcriptional (“T-class”) asymmetry. Each pair of bars (upper axis) shows the density of mutations at C:G (left bar) and G:C (right bar) base pairs. When summing across the entire genome, base-pair orientation does not affect mutational densities. In tx(+) regions, G:C base pairs show a higher density of G→T transversions than C:G base pairs; the opposite is true in tx(-) regions. Lower axis shows the log2 ratio of each pair of bars. (D) POLE-mutant cancers (colorectal and endometrial) show strong replicative (“R-class”) asymmetry. Left-replicating regions show a higher density of mutations at C:G base pairs, and right-replicating regions at G:C. (E) Lung cancers show strong T-class asymmetry but little R-class. (F) POLE-mutant cancers show strong R-class strand asymmetry but little T-class.